Method for separating complete bacteria from infected cells
A complete technology for infecting cells, applied in the direction of isolation of microorganisms, DNA preparation, recombinant DNA technology, etc., can solve the problems of long separation time, large amount of bacterial death, etc., and achieve the effect of shortening the time
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
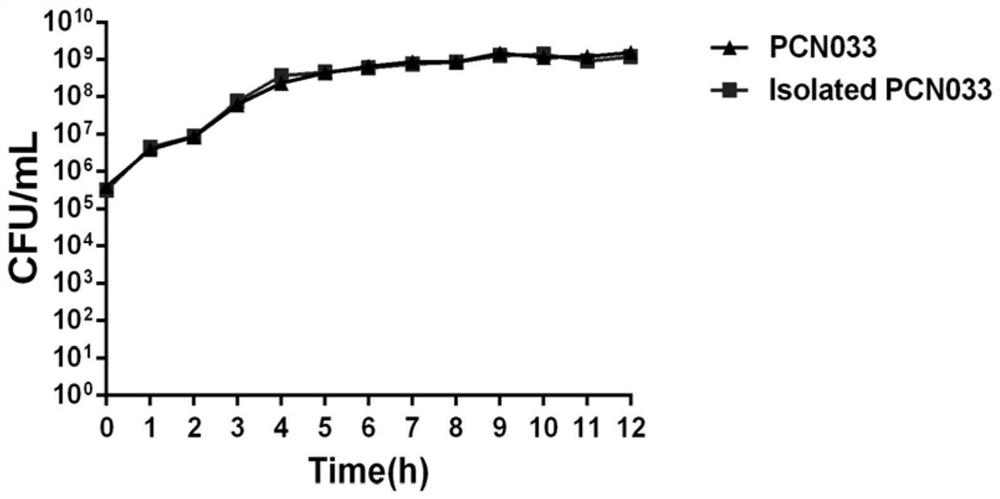
[0021] 1. Bacterial preparation: Take out the frozen porcine parenteral pathogenic Escherichia coli PCN033 strain from -20°C and place it at room temperature. After it dissolves, take 5 μL of the bacterial liquid on the LA solid plate, and use the inoculation silk to Its divisions are demarcated. Then place it in a constant temperature incubator for culture, wait for it to grow a single colony, pick a single colony and culture it in LB medium, after overnight culture, transfer it to fresh LB at a ratio of 1:100 (V / V) The culture is based on shaking culture in a shaker. To be cultured to mid-log phase (OD 600 =0.8), that is, each milliliter contains 5×10 8 When the number of viable cells (CFU) is about 10000000000000000000000000000000000000000000000000000 thallines were obtained, the collected bacteria were resuspended to 0.5 × 10 using RPMI 1640 medium 8 CFU / mL, store in a 4°C refrigerator for later use.
[0022] 2. Bacterial infection of cells: HEp-2 cells frozen at -80°C...
experiment approach 1
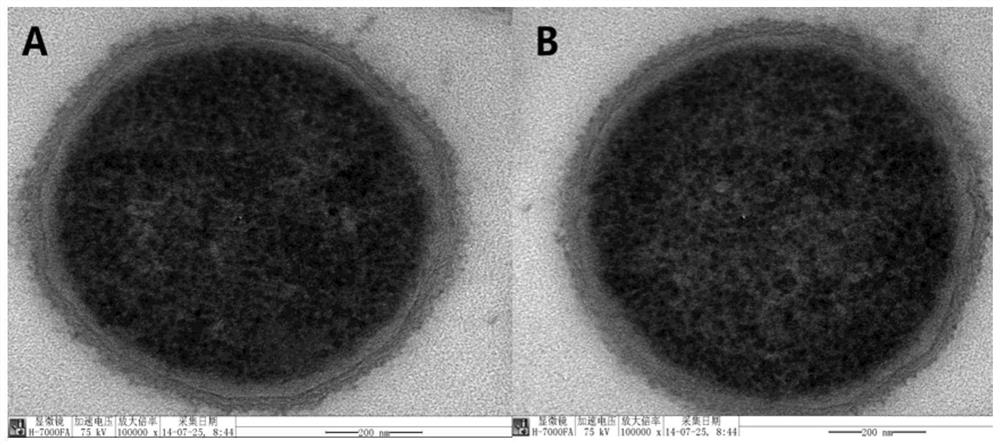
[0025] After adding bacteria to HEp-2 cells, the bacteria were thoroughly contacted with the cells by centrifugation, and then placed in 5% CO 2 constant temperature incubator for cells. After co-incubating at 37°C for 2 hours, discard the medium, add pre-cooled sterile water, and let it stand in the refrigerator at 4°C for 10 minutes, scrape off the adherent cells with a cell scraper, and transfer the cell suspension to a sterile centrifuge. tube, vortexed for 2 minutes, centrifuged at 6000r / min at 4°C for 10 minutes, discarded the supernatant, added pre-cooled PBS, resuspended the pellet, washed three times and collected the bacteria; fixed the collected bacteria with electron microscope fixative , observe the morphological structure of the bacteria through a transmission electron microscope, and measure the growth performance of the isolated and purified bacteria through the growth curve, so as to evaluate the influence of factors such as ice water, cell scraping and vortex...
experiment approach 2
[0027] After adding bacteria to HEp-2 cells, the bacteria were thoroughly contacted with the cells by centrifugation, and then placed in 5% CO 2 constant temperature incubator for cells. After co-incubating at 37°C for 2 h, Triton x-100 at different concentrations (0, 0.01%, 0.025%, 0.05% and 0.1%; V / V) were added, labeled as sample 1, sample 2, sample 3, sample 4 and For sample 5, place it on ice and incubate with shaking, observe the lysis of the cells through an inverted microscope at 0 min, 15 min, 30 min, 60 min and 120 min respectively, and at the same time count the number of viable bacteria in each sample by plate counting method; After complete lysis, transfer the cell lysate containing bacteria to a pre-cooled sterilized centrifuge tube, centrifuge at 6000r / min at 4°C for 10 min, discard the supernatant, add pre-cooled PBS, resuspend the pellet, and wash three times Finally, the bacteria were collected; the collected bacteria were fixed with an electron microscope f...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com