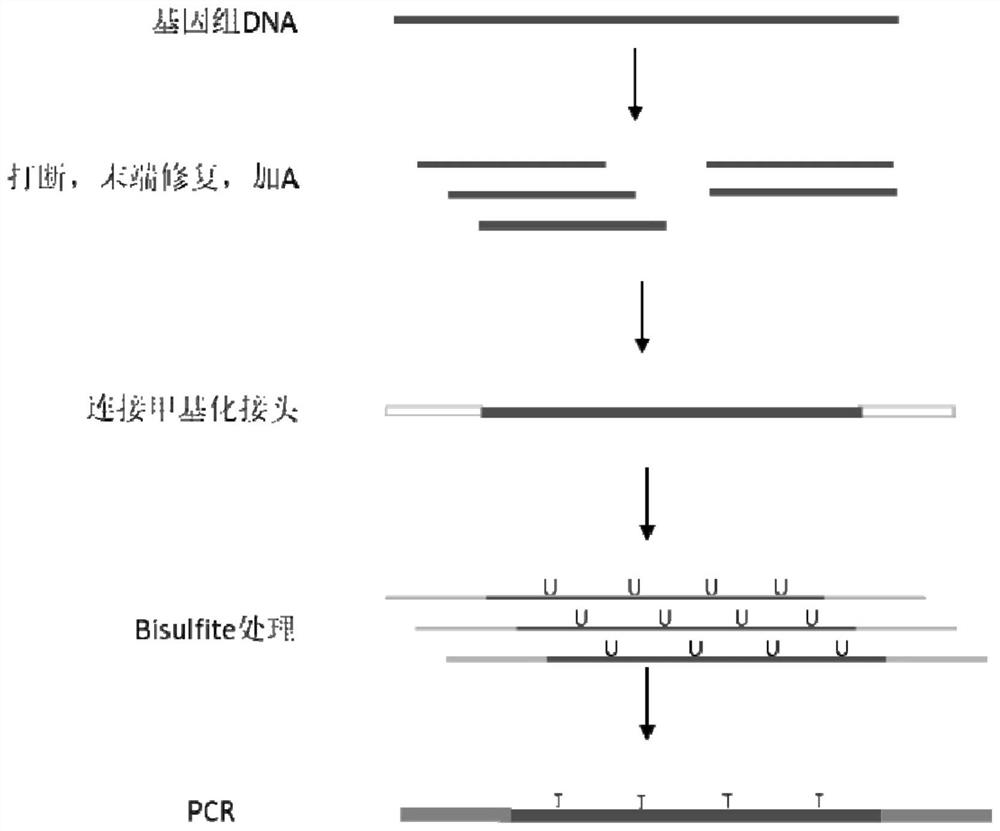
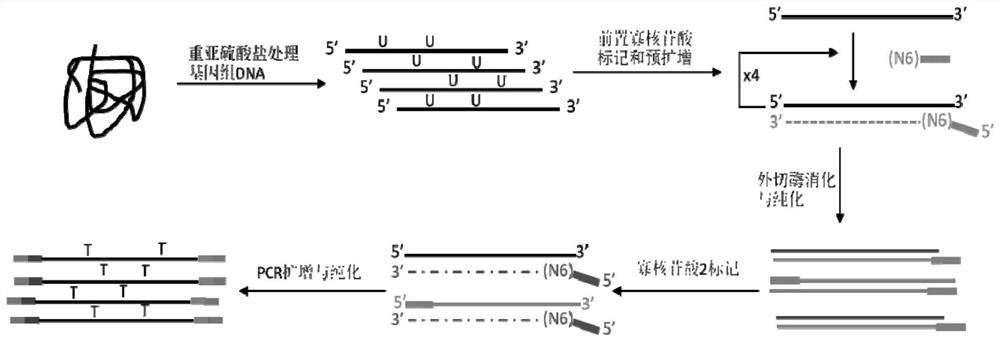
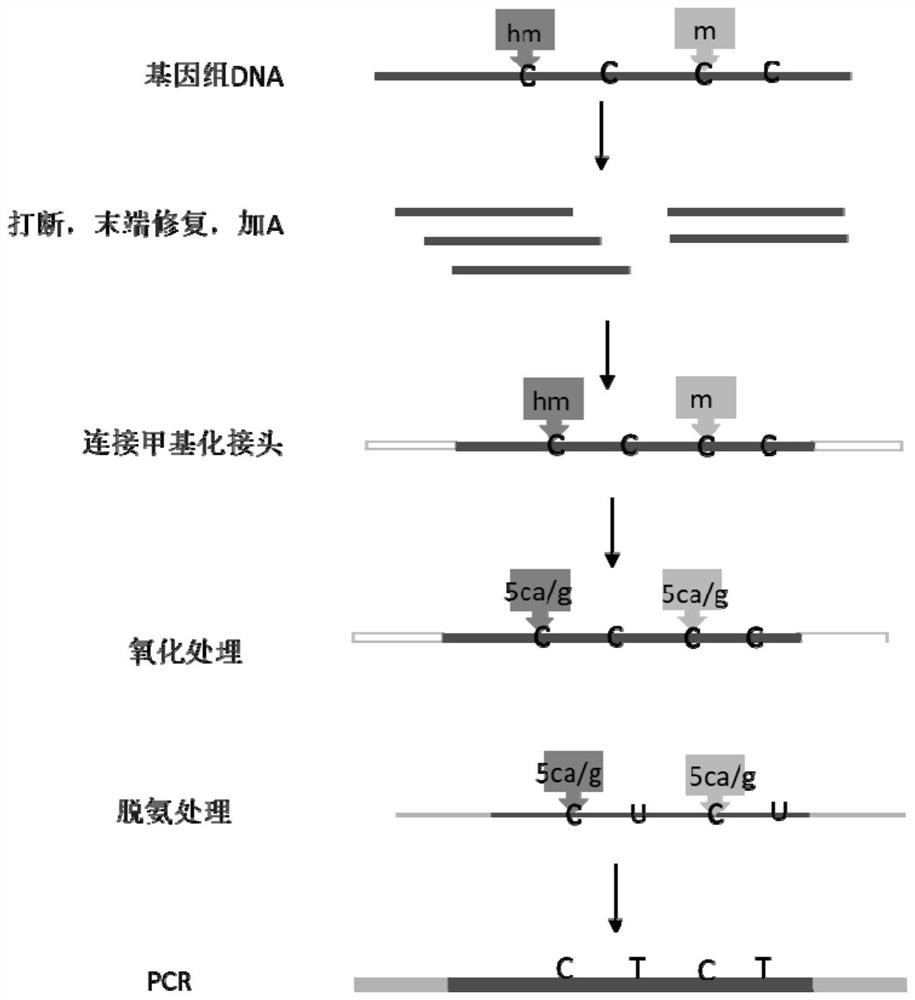
Novel low-initial-quantity DNA methylation library building method
A methylation and sequencing library technology, applied in the field of low-initial DNA methylation library construction, can solve the problems of poor uniformity, low GC coverage, large damage, etc., to improve the unbalanced distribution of GC bases, operation Simple steps and less damage
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0055] Embodiment 1, establishment of method
[0056] 1. Oxidative deamination treatment
[0057] Genomic DNA was taken for oxidative deamination treatment.
[0058] Oxidative deamination method: use EM-seq Conversion Module Kit and operate according to the kit instructions. EM-seq Conversion Module Kit, full name Enzymatic Methyl-seqConversion Module, NEB Corporation, Cat. No. #E7125. Internal control DNA is included in the EM-seq Conversion Module Kit. EM-seq Conversion Module Kit uses TET2 (dioxygenase) for oxidation treatment and APOBEC (cytosine deaminase) for deamination treatment to deaminate unmethylated cytosine C to uracil U.
[0059] 2. Whole Genome Amplification
[0060] The product of step 1 was taken for whole genome amplification.
[0061] Whole-genome amplification method: use REPLI-g Single Cell Kit and perform multiple displacement amplification reactions according to the kit instructions. REPLI-g Single Cell Kit: QIAGEN, Cat. No. 150343; website link...
Embodiment 2
[0072] Embodiment 2, the application of method
[0073] Take Hela cells and extract genomic DNA.
[0074] Take 100pg of genomic DNA and operate according to Example 1.
[0075] Take 10 pg of genomic DNA, and operate according to Example 1.
[0076] The electropherogram of the reaction product of the multiple displacement amplification reaction that completed step 2 is shown in Figure 5 .
[0077] The sequencing results are shown in Table 1.
[0078] Agilent 2100 was used to detect the size of library fragments, the results are shown in Figure 6 (100pg genomic DNA) and Figure 7 (10 pg genomic DNA).
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com