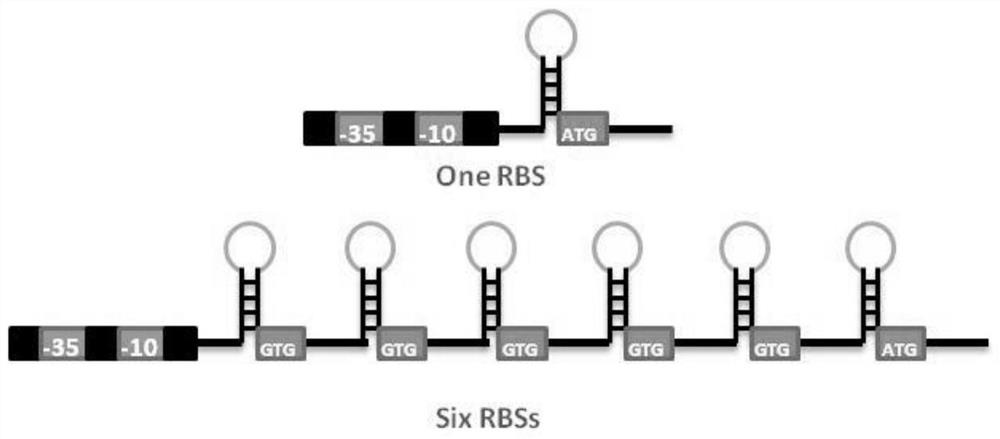
Sequence for increasing translation start sites of gram-positive bacteria and application of sequence in improving protein expression efficiency
A technology of Gram-positive bacteria and starting sites, which is applied in the fields of genetic engineering and microbial engineering, and can solve the problems of insignificant improvement
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0031] The construction of the expression green fluorescent protein (GFP) vector applicable to the 5'-UTR single copy RBS of Bacillus licheniformis and Bacillus subtilis:
[0032] For Bacillus licheniformis and Bacillus subtilis, the gene expression vector is designed based on pHY300PLK:
[0033] 1. According to the Bacillus subtilis (B.subtilis) 168 genome sequence NZ_CP010052.1 released by the NCBI database, find the original sequence of P43, use the Primer Primer 5 software to design primers P43-F and P43-R, and use the total DNA of B.subtilis 168 as The original promoter P43 was amplified from the template, and its 5'-UTR original sequence was GTGATAGCGGTACCATTATAGGTAAGAGAGGAATGTACAC; the primers were designed as follows:
[0034] P43-F: GCGGAATTTCCAATTTCA
[0035] P43-R: GTGTACATTCCTCTCTTACCTATAA
[0036]2. Using the promoter P43 amplified in step 1 as a template, use Primer Primer5 software to design primers P43-F and P43-repeat-R, and change the 5'-UTR region of the P...
Embodiment 2
[0057] Construction of expression green fluorescent protein (GFP) vectors suitable for Bacillus licheniformis and Bacillus subtilis:
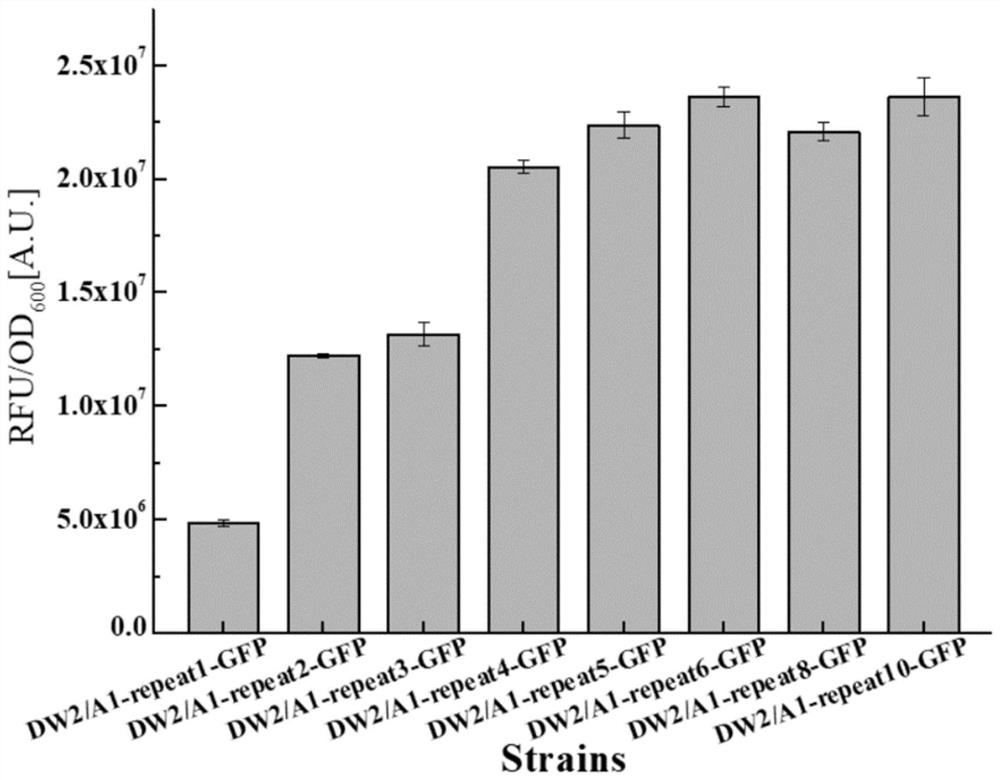
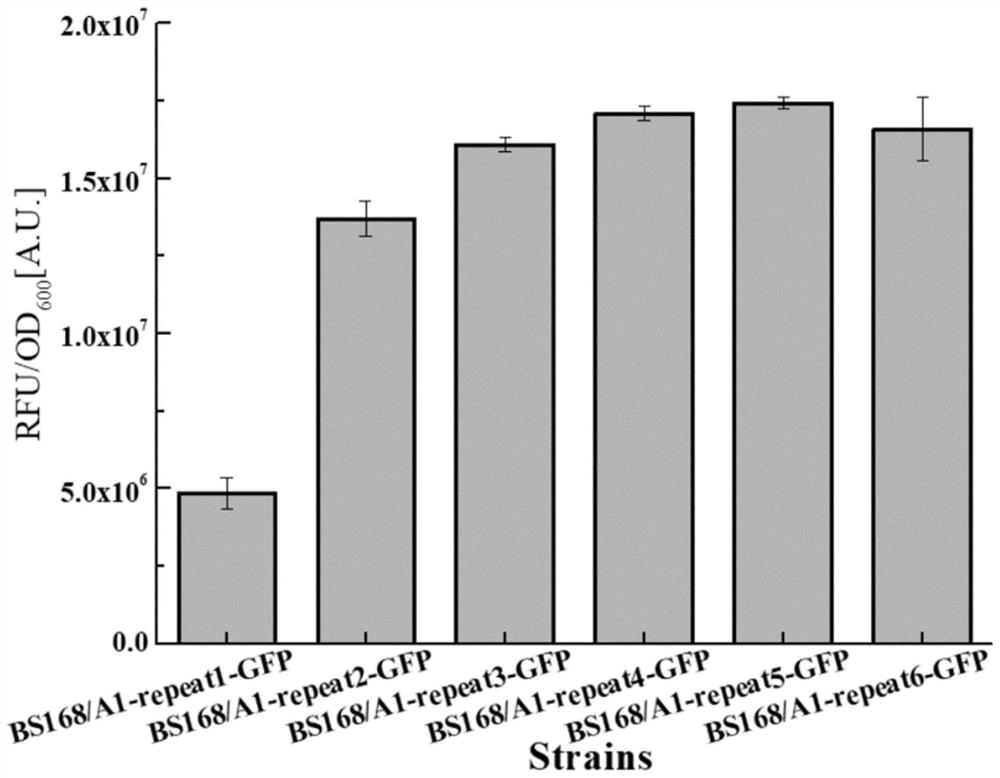
[0058] The multi-copy RBS expression vector is designed with A1-repeat1-GFP:
[0059] 1. Using A1-repeat1-GFP as a template, design primers at the 5'-UTR position. The forward primer GFP-repeat-F contains the 18nt bases at the end of the 5'-UTR of the single-copy plasmid and the 18nt bases at the 5'-end of the GFP gene, and the reverse primer GFP-repeat-R is the reverse primer of two copies of the 5'-UTR. to the sequence;
[0060] Primers were designed as follows:
[0061] GFP-repeat-F: AGAAAGGAGGAAGGATCAATGGTCAGCAAAGGCGAA
[0062] GFP-repeat-R: TGATCCTTCCTCCTTTCTAGATCTGCTCACTGATCCTTCCTCCTTTTCT
[0063] 2. Carry out PCR reaction on the single-copy plasmid A1-repeat1-GFP through the designed primers. The principle of multi-copy amplification is because the GFP-repeat-R primer is a double-copy primer that can bridge from head to tail and cont...
Embodiment 3
[0072] The construction of the expression green fluorescent protein (GFP) vector applicable to the 5'-UTR single copy RBS of Corynebacterium glutamicum 13032:
[0073] For Corynebacterium glutamicum 13032, the gene expression vector was designed based on the commercial vector pEC-XK99E: 1. Using the promoter P of the pEC-XK99E vector trc (excluding the UTR sequence region) and the terminator rrnB T1terminator region are used as the promoter and terminator sequence of this implementation, and the primer pEC-T5-F / R is designed to linearize the vector pEC-XK99E by PCR, and the PCR product is recovered and purified;
[0074] Primers were designed as follows:
[0075] pEC-T5-F:ACACATTATACGAGCCGGAT
[0076] pEC-T5-R: CTGCAGGTCGACTCTAGATTATTTACAGTTCATCCA
[0077] 2. Design the amplification primers for the reporter protein green fluorescent protein (GFP), use the Primer Primer 5 software to design primers GFP-F and GFP-R, and amplify the 5'-UTR single copy RBS green derived from A1...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com