Recombinant protein highly targeting SARS-ConV-2 virus S protein extracellular BD-terminal domain and subunit vaccine thereof
A sars-conv-2, recombinant protein technology, applied in the direction of positive-sense single-stranded RNA viruses, viruses, viral peptides, etc., can solve problems such as breaking the immune balance of the body, achieve good development and application prospects, and improve immune stress ability. Effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
preparation example Construction
[0048] According to another typical implementation of the embodiments of the present invention, a method for preparing a highly targeted recombinant protein of the outer BD end domain of the SARS-ConV-2 virus S protein is provided, the method comprising:
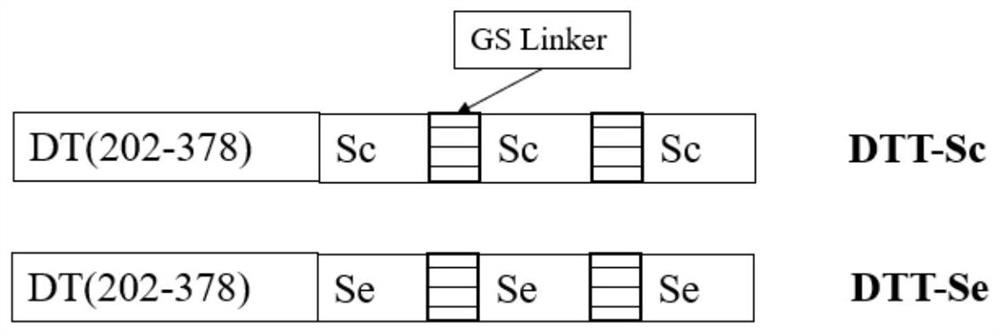
[0049] Obtain the antigenic determinant Sc and Se of SARS-ConV-2 virus S protein membrane extramembrane BD terminal domain domain, the nucleotide sequence of described Sc is as shown in SEQ ID NO: 1, the nucleotide sequence of described Se is as SEQ ID NO: ID NO: 2 shown;
[0050] Constructing and obtaining the expression plasmids of Sc and Se, wherein the nucleotide sequence of the expression region of the Sc expression plasmid is shown in SEQ ID NO: 5, and the nucleotide sequence of the expression region of the Se expression plasmid is shown in Shown in SEQ ID NO: 6;
[0051] The expression plasmids of the Sc and Se were secreted, expressed and purified, respectively, to obtain a highly targeted recombinant protein of the...
Embodiment 1
[0057] Example 1 Analysis and design of epitope in extramembrane domain of BD end of SARS-Cov-2 virus S protein
[0058] 1. Construction of a three-dimensional model of the S protein of the SARA-Cov-2 virus
[0059] Retrieve the amino acid sequences of SARS-Cov-2 and SARS-Cov coronavirus S proteins from the database of the National Center for Bioinformatics (https: / / www.ncbi.nlm.nih.gov), and the retrieval numbers in GenBank are respectively for QHD43416 and P59594. The amino acid sequences of SARS-Cov-2 and SARS-Cov S proteins were compared by Clustal software, and their amino acid identity was 76%, showing high homology. The highly homologous sequences of the S proteins of the two viruses are mainly concentrated in the middle to the C-terminus. A three-dimensional structural model of the 2019-nCoV viral S protein was further constructed. The 2019-nCoV virus forms the spike structure on the surface of the virion envelope through the polymerization of three S proteins. Fur...
Embodiment 2
[0074] Example 2 Expression and Purification of S Protein BD-related Epitopic Determinant Peptides
[0075] 1. Expression of 2Sc and 3Se
[0076] The constructed plasmids were respectively transformed into BL21(DE3) expression competent cells, coated on LB plates containing Kanna resistance, placed in a 37°C incubator, and cultured overnight to grow single clones. Four single clones were selected respectively, and cultured in 5 ml of Kanna-resistant LB medium at 200 rpm at 37° C. for 5 h. When the OD600 reached 0.6-0.8, take out 500 μL and put it into a new 1.5mL EP tube marked as before induction. The remaining bacterial liquid was induced by adding IPTG (final concentration 1 mM), and the samples marked as pre-induction and post-induction samples were placed at 37° C. and incubated for 4 hours. Aspirate 200 μL of the bacterial solution before and after induction, centrifuge at 12000 rpm for 5 min. Remove the supernatant, add 50 μL of PBS to resuspend, add 2*loading buffer...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com