Recombinant corynebacterium having 1,3-pdo productivity and reduced 3-hp productivity, and method for producing 1,3-pdo by using same
A 3-PDO, microbial technology, applied in the direction of microbial-based methods, biochemical equipment and methods, chemical instruments and methods, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
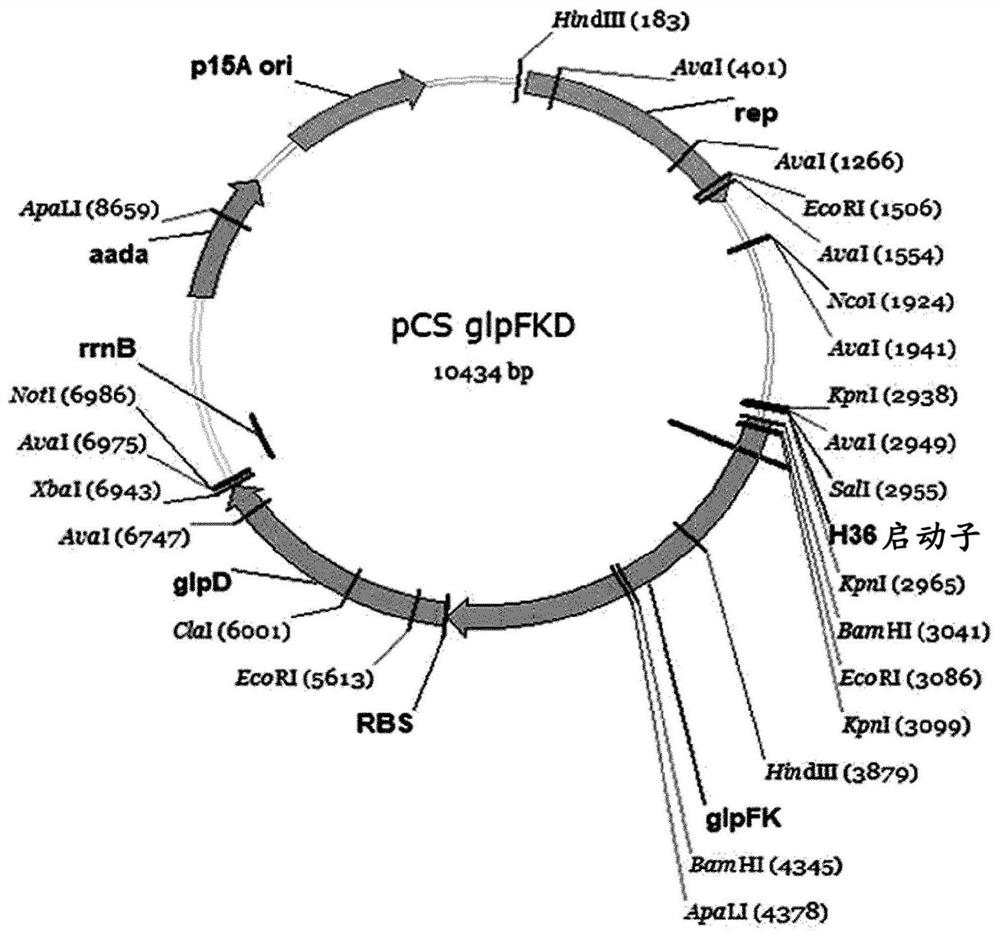
[0048] In the following examples, only genes derived from specific strains are given as examples of genes to be introduced, but it will be apparent to those skilled in the art that any genes can be used without limitation as long as they are used in the genes to be introduced into them. It is enough to express in the host and exhibit the same activity. Example 1: Generation of the pCSglpFKD vector for the production of mutant C. glutamicum capable of growth using glycerol as a single carbon source
[0049] 1-1: Production of pCSglpFKD vector for constructing glycerol catabolism pathway
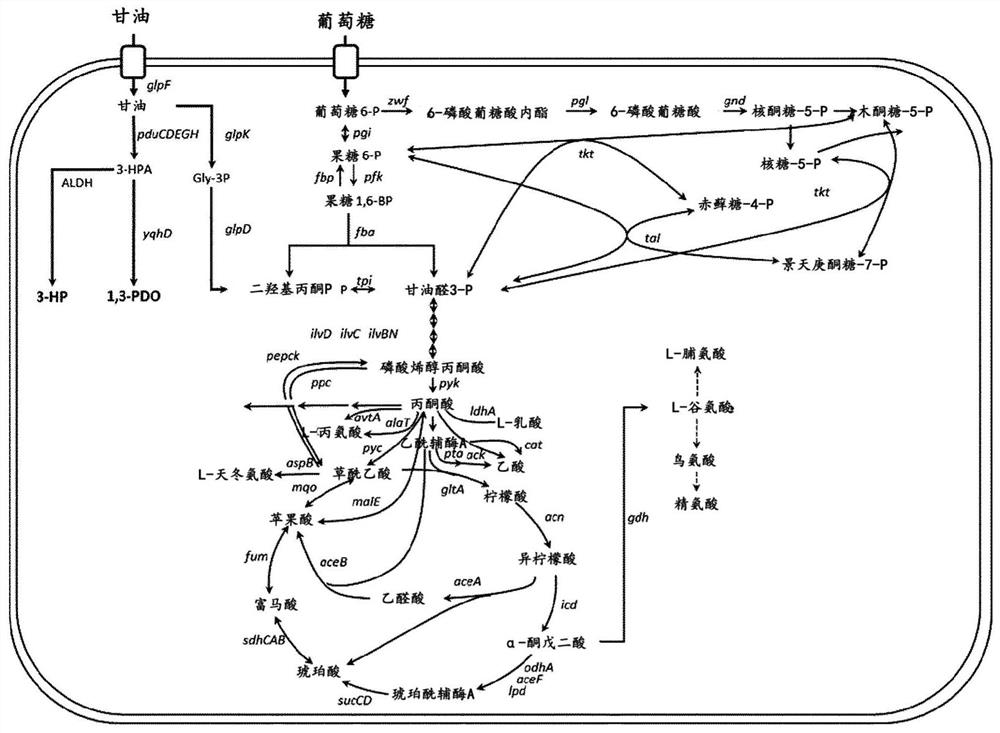
[0050] C. glutamicum is known to be unable to use glycerol as a single carbon source for cell growth. Therefore, to construct a glycerol catabolism pathway, a gene encoding an enzyme derived from Escherichia coli W3110 and responsible for a glycerol catabolism pathway was first expressed using the Corynebacterium glutamicum shuttle vector pCES208s-H36-S3.
[0051] Use the chromosomal DNA of ...
Embodiment 2
[0056] Example 2: Production of aldehyde dehydrogenase deletion vectors for inhibition of 3-HP biosynthesis
[0057] When 1,3-PDO is produced from glycerol, the precursor 3-HPA produced is converted to 3-HP by aldehyde dehydrogenase present in the cell. However, an enzyme catalyzing a reaction accepting a precursor as a substrate has not been reported in C. glutamicum. Therefore, in order to identify the aldehyde dehydrogenase that mediates the reaction and delete the gene encoding the enzyme from the genome of the strain to thereby inhibit 3-HP biosynthesis, the 13 aldehyde dehydrogenases present in Corynebacterium glutamicum were first selected. Hydrogenase (Table 2).
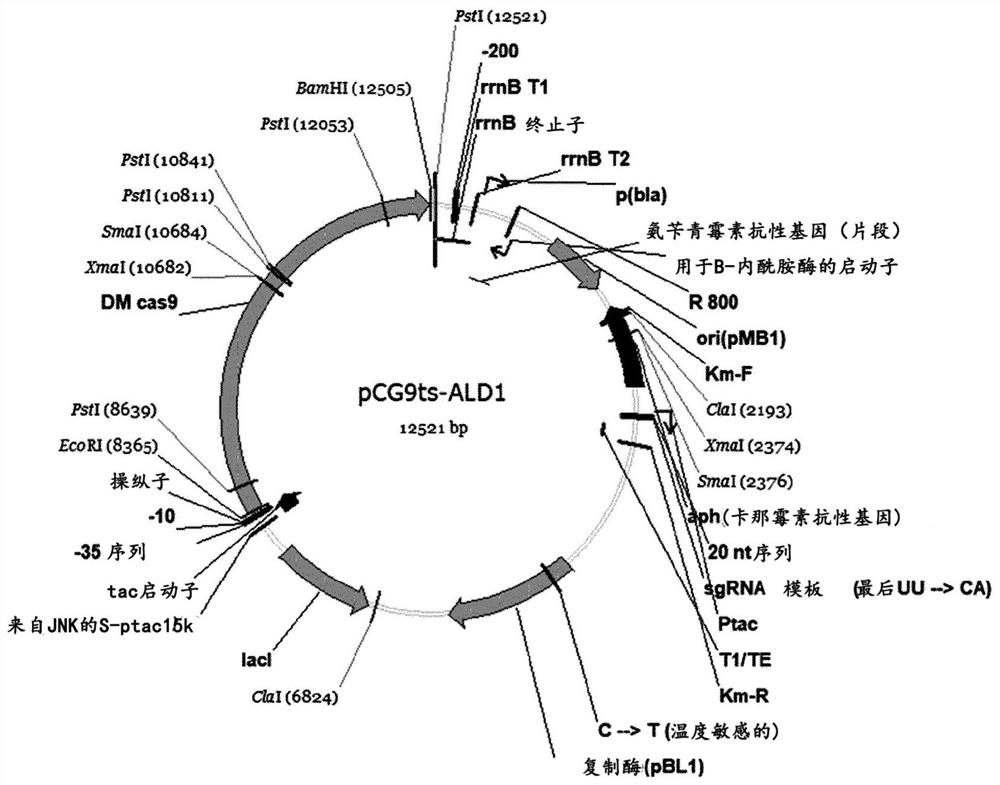
[0058] Then, in order to confirm the inhibitory effect on 3-HP biosynthesis by deleting the genes (SEQ ID NO: 56 to 68) encoding these 13 aldehyde dehydrogenases, pTacCC1- Strains of the HrT vector (Cho et al., Metabolic Engineering, 42:157-167, 2017). Then, i) pCG9ts series each containing sgRNA sequences...
Embodiment 3
[0078] Example 3: Production and confirmation of Corynebacterium glutamicum with suppressed 3-HP production capacity and improved 1,3-PDO production capacity
[0079] 3-1: Production of Corynebacterium glutamicum having suppressed 3-HP production ability
[0080] The pCG9ts-ALD vector and ssODN produced in Examples 2-1 and 2-2 were each transformed into wild-type Corynebacterium glutamicum (ATCC 13032) in order to delete from the genome any coding sequence expected to be involved in 3-HP biosynthesis. Genes for 13 aldehyde dehydrogenases. Then, for the transformed mutant Corynebacterium glutamicum strain, the pTacCC1-HrT vector was removed by solidification on a 37°C BHI plate (Korean Patent Application No. 2017-0042124; Cho et al., Metabolic Engineering, 42:157-167, 201, SEQ ID NO:57) and pCG9ts-ALD vector. The strains produced by this process are shown in Table 6. However, the WAH3 and WAH9 strains were not produced, and the corresponding two genes were considered essenti...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com