Bacterial HI-C genome and plasmid conformation capturing method
A HI-C and genomic technology, applied in biochemical equipment and methods, combinatorial chemistry, recombinant DNA technology, etc., can solve problems such as error-prone and immature, and achieve highly practical results
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
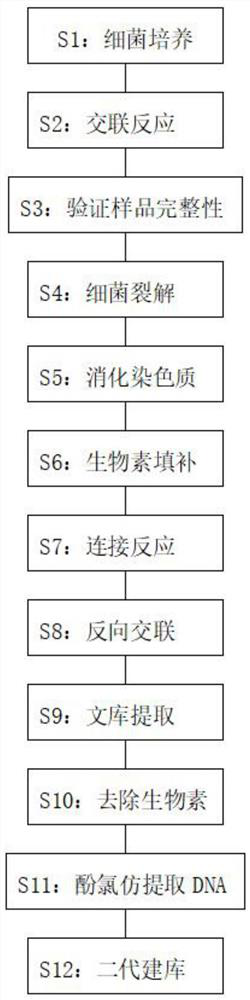
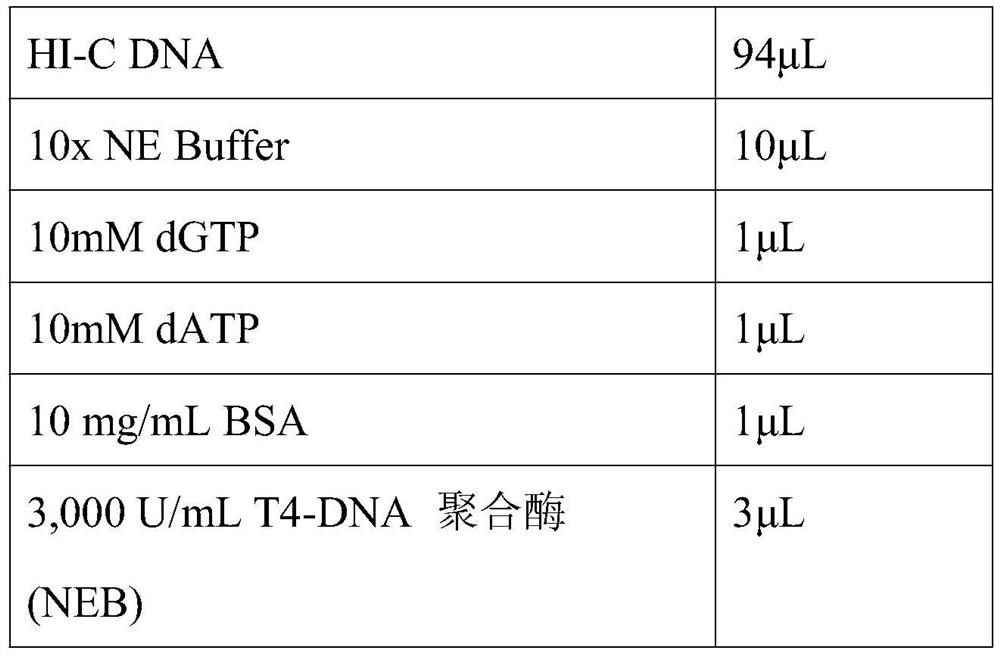
Embodiment
[0040] The first step: bacterial culture.
[0041] 1. Dip a small amount of bacterial solution with an inoculation loop, streak it on an LB agar plate (or an identification agar plate), and place the plate in a 37°C incubator for 18-24 hours.
[0042] 2. Cultivate the single bacterium in 50 mL of LB broth at 37°C on a shaker for 3 hours and 15 minutes.
[0043] 3. Collect all the bacterial liquid, centrifuge at 5000 rpm at 4°C for 8 minutes, discard the culture liquid, and collect the bacterial precipitate.
[0044] 4. Add 6mL of fresh LB broth, resuspend the bacteria, and wash. Then, centrifuge at 5000 rpm for 8 minutes at 4°C to collect the bacterial pellet.
[0045] The second step: cross-linking reaction.
[0046] 1. Prepare 20mL of 3% formaldehyde solution (protected from light on ice): 3% formaldehyde solution
[0047] 18.4mL 1*PBS solution + 1.6 / mL 37% w / v formaldehyde solution
[0048] 2. Add the prepared 3% formaldehyde solution into a 50mL centrifuge tube contai...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com