Chrysanthemum chloroplast genomic SSR marker library, acquisition method and application thereof
A chloroplast genome and genome technology, applied in the field of molecular biology, can solve the problems of hindering variety exchange and cultivation, low identification efficiency, confusing chrysanthemum varieties, etc., to avoid chloroplast DNA isolation, high-efficiency expression and safety, and improve system classification efficiency Effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
[0030] Below, in conjunction with accompanying drawing and specific embodiment, the present invention is described further:
[0031] Genome comparative analysis and synteny analysis
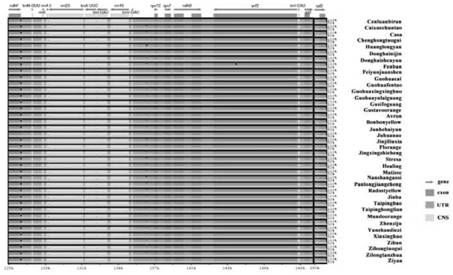
[0032] Taking the annotation information of the SSR marker sequence SEQ ID NO.1 of Chrysanthemum Chrysanthemum indicum (JN867589.1) as a reference, use mVISTA (Frazer et al., 2004) to select the alignment method of Shuffle-LAGAN, for the 39 The SSR marker sequence SEQ ID NO.2-40 of three chrysanthemum varieties was compared and analyzed; the SSR marker sequence collinearity analysis was performed on the basis of the chloroplast genome by the Genious10.0.9 Mauve tool (Darling et al., 2004).
[0033] Table 1 Chrysanthemum Varieties
[0034]
[0035]
[0036] Repeat sequence and SSR analysis
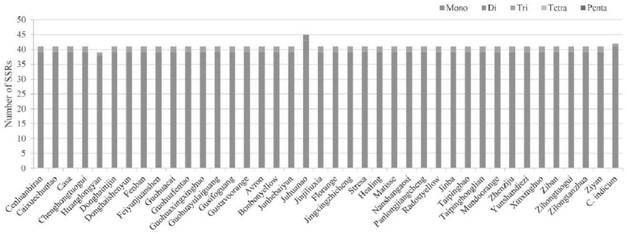
[0037]Use MISA's perl script (Beier et al., 2017) for simple sequence repeats (simple sequence repeats, SSR) analysis, the minimum number of repeats from single nucleotide to hexanucleotide units is se...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com