Polypeptide for inhibiting novel coronavirus (SARS-COV-2) and application of polypeptide
A virus and virus infection technology, applied in the direction of viral peptides, viruses, antiviral agents, etc., to prevent membrane fusion and prevent viruses from infecting cells
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0035] Search for HR area:
[0036] Download the protein sequence of the S protein of SARS-COV-2 from the NCBI database (GenBank: QHQ82464.1) and perform multiple sequence alignment with the HR1 and HR2 sequences of SARS and MERS respectively through ClustalW software (the gap penalty is set to 10 , the extension penalty is set to 0.2). The HR1 and HR2 sequences of SARS and MERS will be aligned to SARS-COV-2, and the aligned SARS-COV-2 sequence will be its HR region.
[0037] The sequence of HR1 is shown as amino acid in SEQ ID NO:1:
[0038]YRFNGIGVTQNVLYENQKLIANQFNSAIGKIQDSLSSTASALGKLQDVVNQNAQALNTLVKQLSSNFGAISSVLNDILSRLDKVE
[0039] The sequence of HR2 is shown as amino acid in SEQ ID NO:2:
[0040] ELDKYFKNHTSPDVDLGDISGINASVVNIQKEIDRLNEVAKNLNESLIDLQELGK YEQYIKWPWY
Embodiment 2
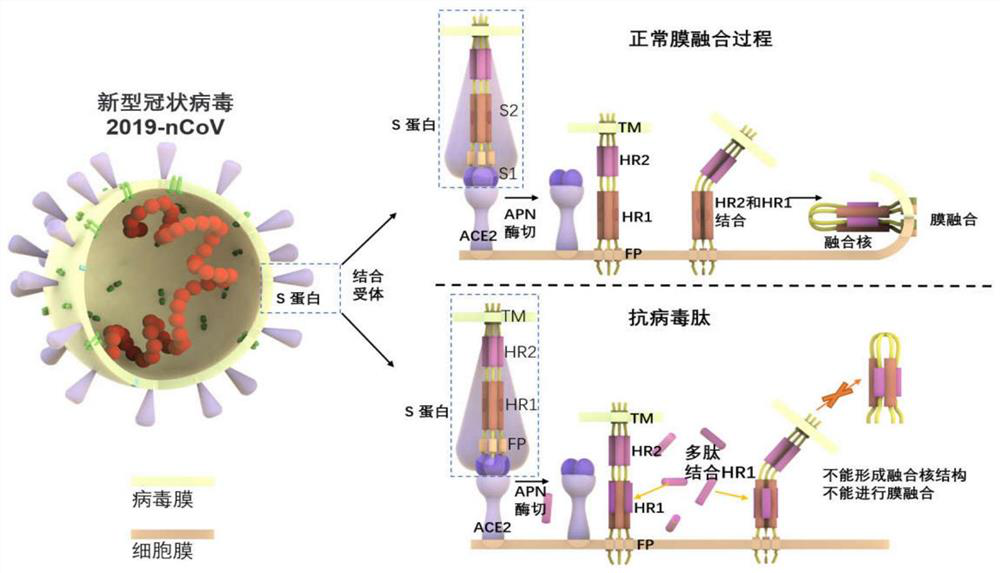
[0042] 1) Modeling of HR1 / HR2 fusion nucleus:
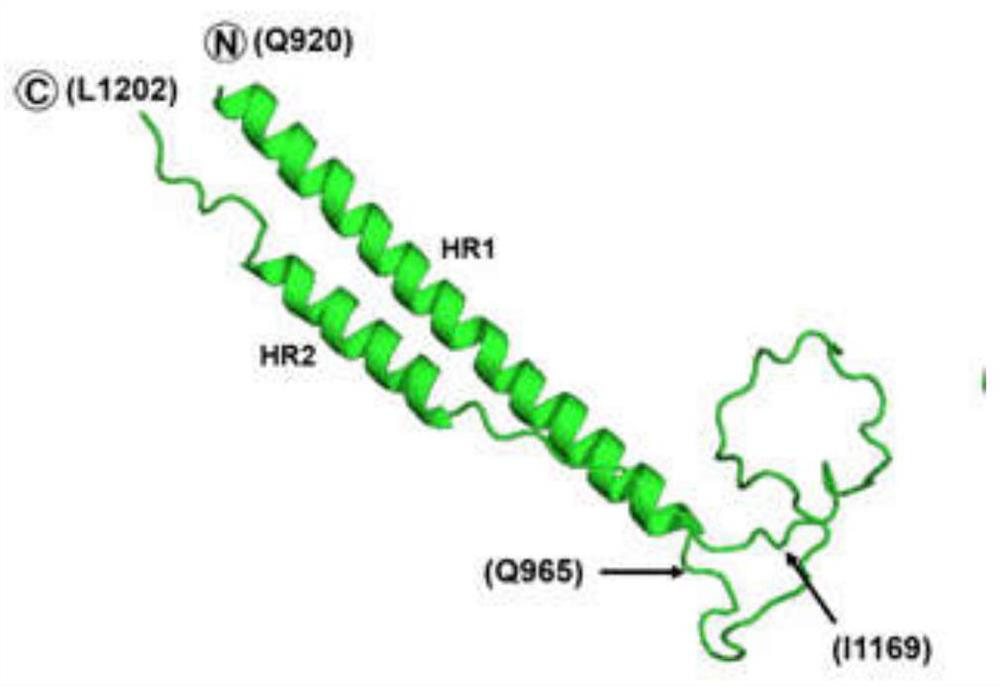
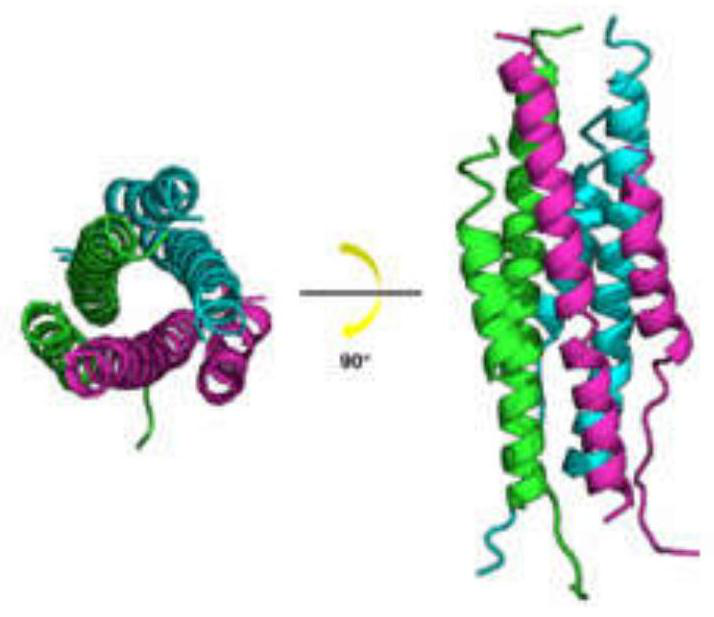
[0043] Using the HR region obtained above, place HR1 at the N-terminal of the complex, place HR2 at the C-terminal of the complex, and connect HR1 and HR2 with a linking sequence of 22 amino acids according to conventional techniques to obtain HR1 / HR2 fusion core; wherein, the linking sequence is LVPRGSGGSGGSGGLEVLFQGP (see figure 2 ).
[0044] Depend on figure 2 The overall structure of the HR1 / HR2 fusion core can be seen, and the two helix structures in the molecule correspond to the HR1 and HR2 domains. The HR1 domain ranges from Q920 to Q965 and the HR2 domain ranges from I1163 to L1202. The fragment between Q965 and I1169 is the connecting peptide. It can be observed that HR2 is mainly connected with two HR1 fragments in the HR1 / HR2 trimer through hydrogen bonds and hydrophobic interactions ( image 3 B).
[0045] After obtaining the HR1 / HR2 complex, use the protein sequence of the complex to model: use the SWISS-MO...
Embodiment 4
[0060] The inhibitory peptides obtained above and the modified inhibitory peptides were tested for their ability to bind to proteins by molecular docking and molecular dynamics simulations:
[0061] 1) Experimental method:
[0062] Use PyMOL to delete the HR1 and connecting peptide in the structure, and separate out the individual HR2 fragments. After HR2 is isolated from the fusion nucleus, use the GRAMM-X website (http: / / vakser.compbio.ku.edu / resources / gramm / grammx / ) to dock HR2 or the above-mentioned examples to obtain inhibitory peptides or modified peptides to the fusion nucleus. Molecular simulations were then performed using GROMACS software. Set the umbrella sampling window to 0.4, and take the 170 to 330 frames of the simulation results for umbrella sampling. Use the wham command to calculate its energy through the pullf.xvg file in the result (use the WeightedHistogram Analysis Method (WHAM) to calculate the Potential of Mean Force (PMF)). The analysis process is...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com