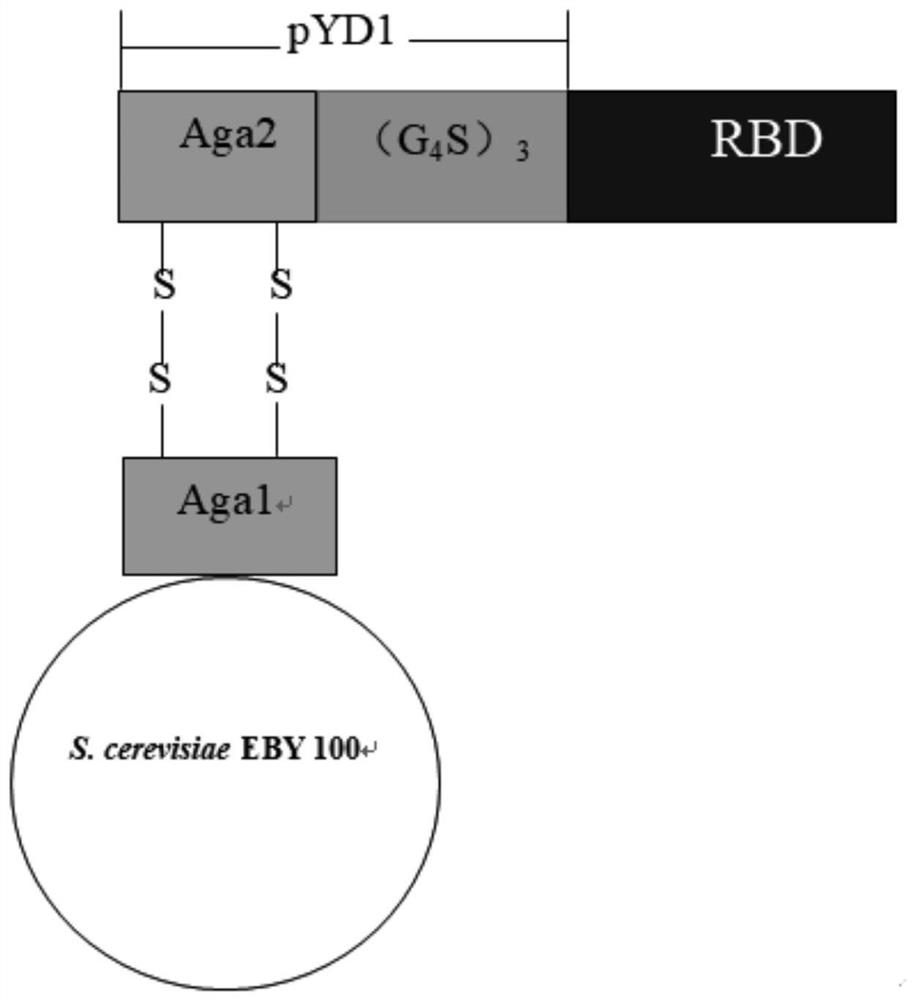
COVID-19 subunit vaccine based on saccharomyces cerevisiae surface display system
A technology of yeast and recombinant yeast, which is applied in the field of antiviral vaccines, can solve the problems of vaccine safety key protein concept stability and immune response level to be further confirmed, and achieve the effect of improving immunity and preventing infection
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
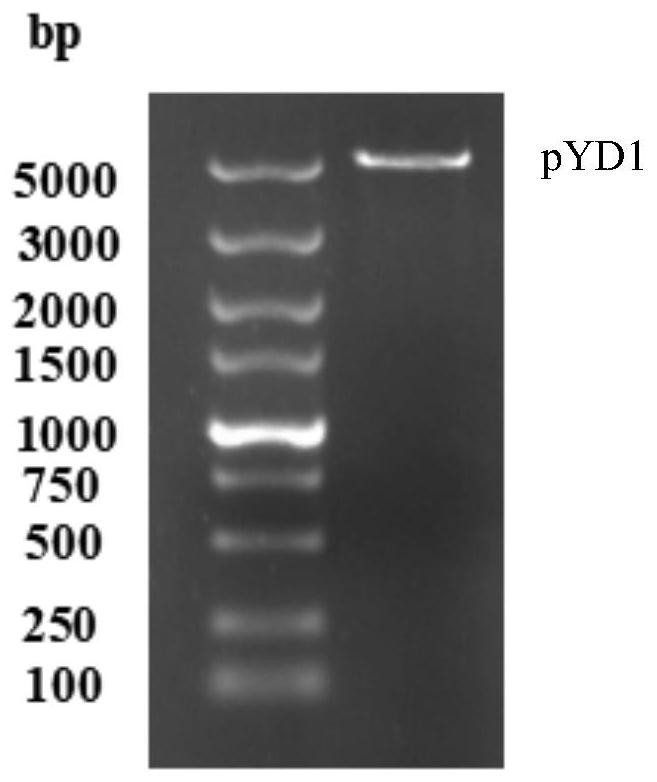
[0080] The construction of embodiment 1 recombinant plasmid pYD1-RBD
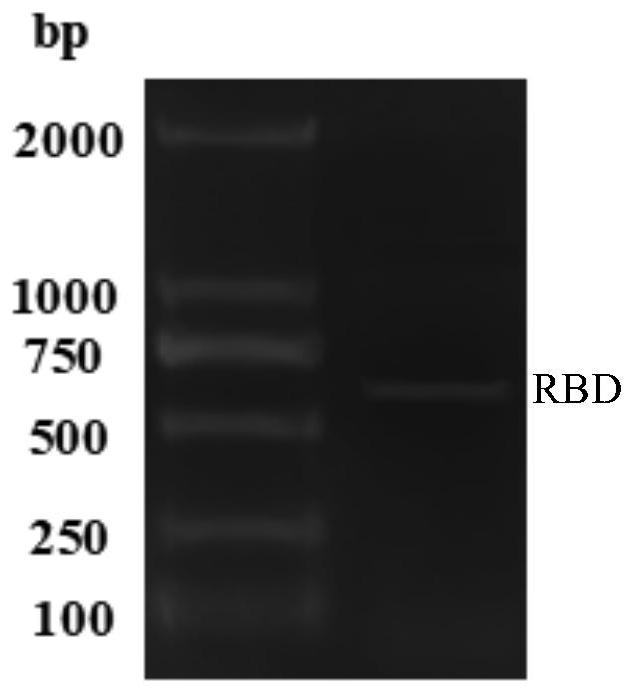
[0081] 1. RBD gene amplification
[0082] 1.1 Design of RBD-specific primers
[0083] Using the S gene of SARS-CoV-2 (gene bank number: MN908947) as a DNA template, the RBD gene was amplified by designing specific primers. The primer sequences are shown in Table 1, and the underlines in primers F-1 and R-1 are respectively Restriction sites Nhe I and EcoR I.
[0084] Table 1. PCR primers for RBD gene
[0085]
[0086] The receptor binding domain (RBD) gene sequence (SEQ ID NO.1, 582bp) is as follows:
[0087] ATGCCTAATATTACAAACTTGTGCCCTTTTGGTGAAGTTTTTAACGC CACCAGATTTGCATCTGTTTATGCTTGGAACAGGAAGAGAATCAGCAAC TGTGTTGCTGATTATTCTGTCCTATATAATTCCGCATCATTTTCCACTTTTA AGTGTTATGGAGTGTCTCCTACTAAATTAAATGATCTCTGCTTTACTAAT GTCTATGCAGATTCATTTGTAATTAGAGGTGATGAAGTCAGACAAATCG CTCCAGGGCAAACTGGAAAGATTGCTGATTATAATTATAAATTACCAGAT GATTTTACAGGCTGCGTTATAGCTTGGAATTCTAACAATCTTGATTCTAA GGTTGGTGGTAATTATAATTACCTGTATAGATTGTTTAGGAAGTC...
Embodiment 2
[0123] Example 2, Molecular Construction of Recombinant Yeast EBY100 / pYD1-RBD
[0124] 1. Transformation of recombinant plasmid pYD1-RBD
[0125] (1) Take the EBY100 strain stored in a -80°C ultra-low temperature refrigerator, streak it on the YPD solid medium, place it in an incubator, and cultivate it at 30°C;
[0126] (2) Pick a single colony in 3mL YPD medium and culture at 30°C;
[0127] (3) Measure the absorbance (OD) of EBY100, and transfer the cultured bacterial liquid into 50 mL of fresh YPD culture liquid, adjust the OD value to 0.4, and continue to cultivate for 4-5 hours at 30°C and 200 rpm;
[0128] (4) Centrifuge at 3,000 rpm for 5 minutes at 4°C, wash twice with sterile water;
[0129] (5) Centrifuge at 3,000 rpm for 5 minutes at 4°C, and resuspend the bacteria in 1 mL of 0.1M LiAc;
[0130] (6) Put it on the prepared ice for 10 minutes, centrifuge at 4,000 rpm for 3 minutes, discard the supernatant, resuspend the pellet in 300 μL LiAc, take 2 EP tubes, and d...
Embodiment 3
[0143] Embodiment 3, the preparation of oral vaccine
[0144] (1) Induce expression of recombinant Saccharomyces cerevisiae EBY100 / pYD1-RBD.
[0145] (2) Take the bacterial liquid induced for 72 hours, and measure its absorbance at OD600nm.
[0146] (3) Centrifuge at 5,000 rpm for 15 min at 4°C, discard the supernatant, and rinse the pellet three times with sterilized PBS.
[0147] (4) The bacterium was resuspended in sterilized PBS, and its concentration was adjusted to 1OD600nm / μL, and inactivated in a water bath at 60°C for 1 hour.
[0148] The induction expression method in step (1) is as follows:
[0149] (a) A single colony of EBY100 / pYD1-RBD prepared in Example 2 was picked with a pipette gun, inoculated into 3 mL of culture medium, placed in a vibrating shaker, and cultured at 30° C. and 250 rpm for 16-18 hours.
[0150] (b) Take 1 mL of bacterial liquid to measure the OD600nm value, so that the OD600nm value is between 2.0-5.0.
[0151] (c) In the YNB-CAA liquid m...
PUM
| Property | Measurement | Unit |
|---|---|---|
| molecular weight | aaaaa | aaaaa |
| adsorption capacity | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com