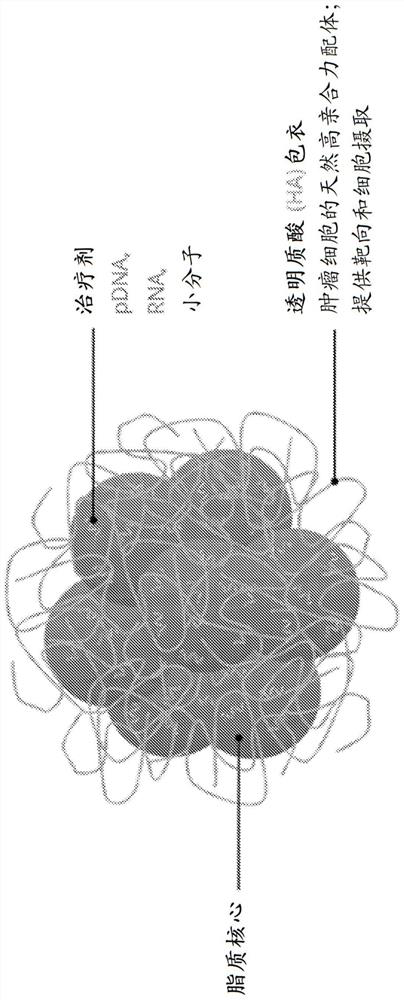
Encapsulated polynucleotides and methods of use
A polynucleotide, ribozyme technology, used in immunology, inflammation and cancer therapy, to solve the problems of peripheral cell and tissue disease collateral damage, limit the application of viral therapeutics, hinder the efficacy of repeated systemic administration, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0255] Example 1: Engineering of Polynucleotide Constructs Encoding Replication Competent Viral Genomes
[0256] The autonomously replicating polynucleotide constructs described herein are engineered and generated using standard molecular biology and genetics techniques. Exemplary constructs encoding specific viruses and the corresponding cancers treated with these constructs are described in Tables 13, 14 and 15 below. However, an appropriate virus can be selected based on the desired properties of the virus and the properties of the cancer to be treated. Similarly, miRNA target sequence cassettes (miR TS) can be inserted at one or more locations in the viral genome to control replication of the encoded viral genome in normal non-cancerous cells while allowing replication in cancerous cells. Exemplary constructs are described throughout this disclosure. The constructed constructs are summarized in Table 8 below.
[0257] Table 8: Polynucleotide constructs encoding replicat...
Embodiment 2
[0260] Example 2: Design and Production of Plasmids Comprising Polynucleotide Constructs Encoding Replication Competent Viral Genomes
[0261] Poly(A), 5'hammerhead ribozyme and 3'delta hepatitis ribozyme were added by fusion PCR after SVV viral DNA was synthesized in Genscript and inserted into the base vector using Gibson assembly. The base vector is 2.4 kb in length and contains a minimal origin of replication and a kanamycin resistance cassette that has been optimized for use in mammalian cells ( Figure 31A ). The expression cassette is disclosed as SEQ ID NO:1. A similar vector was constructed against Coxsackievirus (CVA21) and was Figure 31B shown in . The CVA21 expression cassette is disclosed as SEQ ID NO:2.
Embodiment 3
[0262] Example 3: Design and production of NanoV constructs flanked by ITRs
[0263] To generate ITR-flanked NanoV constructs, the autonomously replicating polynucleotide construct was inserted into an expression cassette flanked by AAV-derived ITRs under the control of a tetracycline (Tet)-responsive promoter. Figure 17 A schematic diagram of the model NanoV construct is provided. The tetracycline-responsive promoter (TRE Tight) drives the expression of mCherry, which serves as a placeholder and can be replaced by an appropriate viral genome construct ( Figure 17 shown as OV). Tetracycline-controlled transactivator (tTA) expression is controlled by a constitutive promoter, in Figure 17Shown as UbCP in . The NanoV construct was inserted into the UL3 / 4 intergenic region of HSV-1 using the Gateway cloning system (Thermo Fisher) which allows rapid insertion of different NanoV cassettes. Addition of tetracycline to the medium causes Tet to bind to tTA, preventing expression...
PUM
| Property | Measurement | Unit |
|---|---|---|
| Average size | aaaaa | aaaaa |
| The average diameter | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com