Linear probe and method for detecting miRNA by using same
A probe detection, linear technology, applied in the direction of DNA / RNA fragments, recombinant DNA technology, biochemical equipment and methods, etc., can solve the problems of insufficient sensitivity or specificity, long reverse transcription time, etc., to reduce steric hindrance, Fast response and good detection specificity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
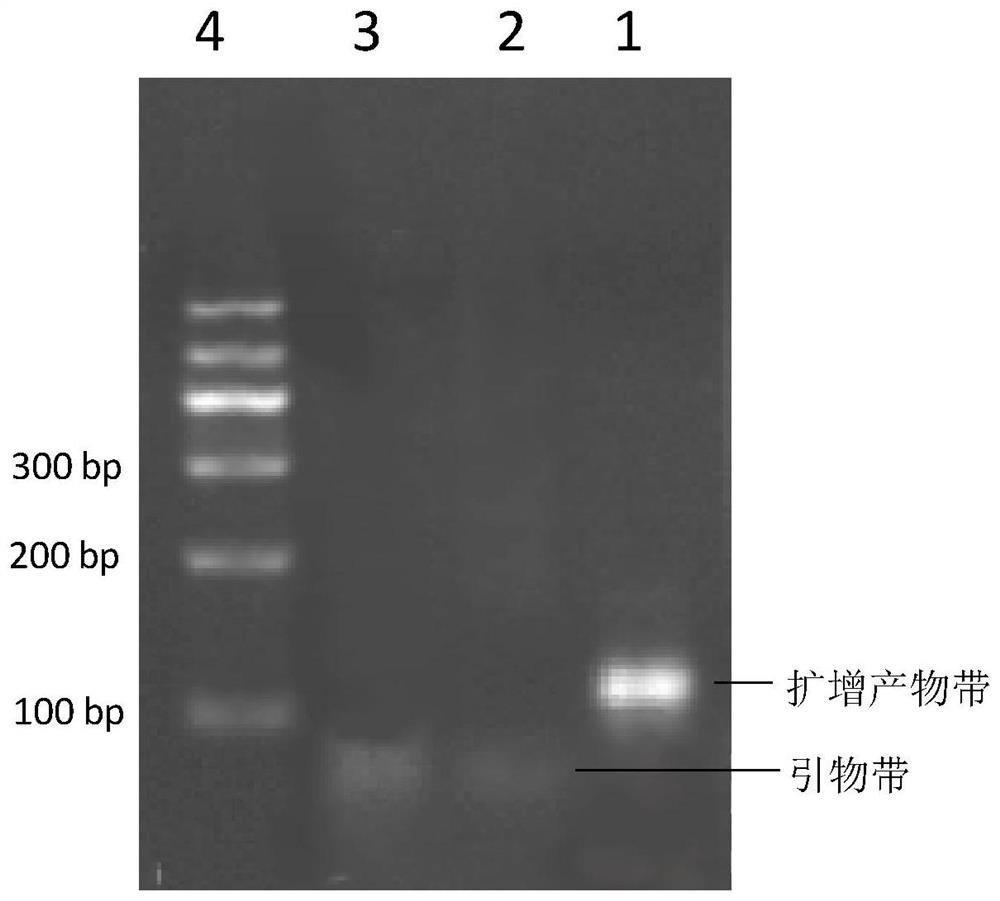
[0050] The feasibility of embodiment 1 verification method
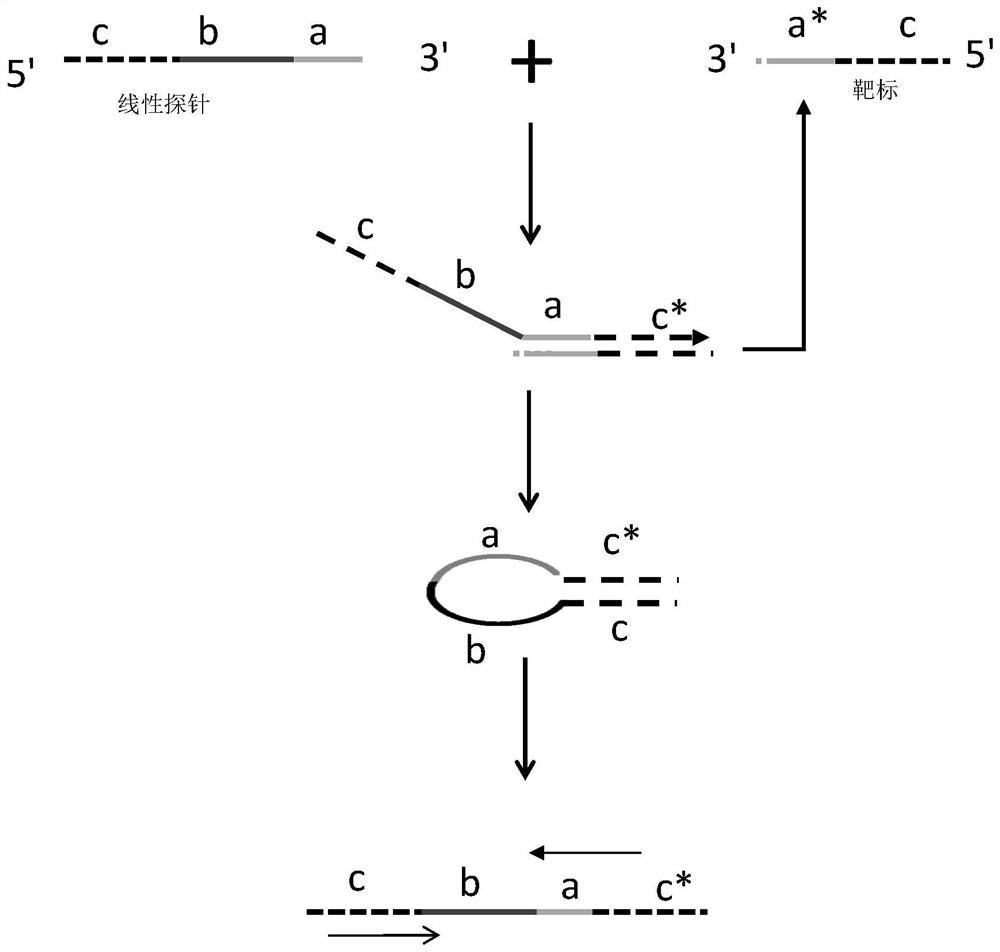
[0051] The linear probe of the present invention includes three regions a, b and c. The 3' end of the b region is connected to the 5' end of the a region, the 3' end of the c region is connected to the 5' end of the b region; the nucleotide sequence of the a region is complementary to the target nucleic acid, and the b region The nucleotide sequence assists loop formation by reducing steric hindrance, and the nucleotide sequence of the c region is the same as the 5' end of the target nucleic acid. In this example, miRNA155 is used as the target miRNA (sequence: UUAAUGCUAAUUGUGAUAGGGGU, ie, SEQ ID NO.1), and miRNA155 linear probe 1 (sequence: TTAATGCTAATTGGCATGAAACCAGTGCTGAGTGTCAGAGACCCCTATCAC, ie, SEQ ID NO.2, a, b, c of the linear probe is designed) See Table 1 for the three regions), PCR-F1 (sequence: TTAATGCTAATTGGCATGAAAC, ie SEQ ID NO.3) and PCR-R1 (sequence: TTAATGCTAATTGTGATAGGGGT, ie SEQ ID NO.4) for reverse...
Embodiment 2
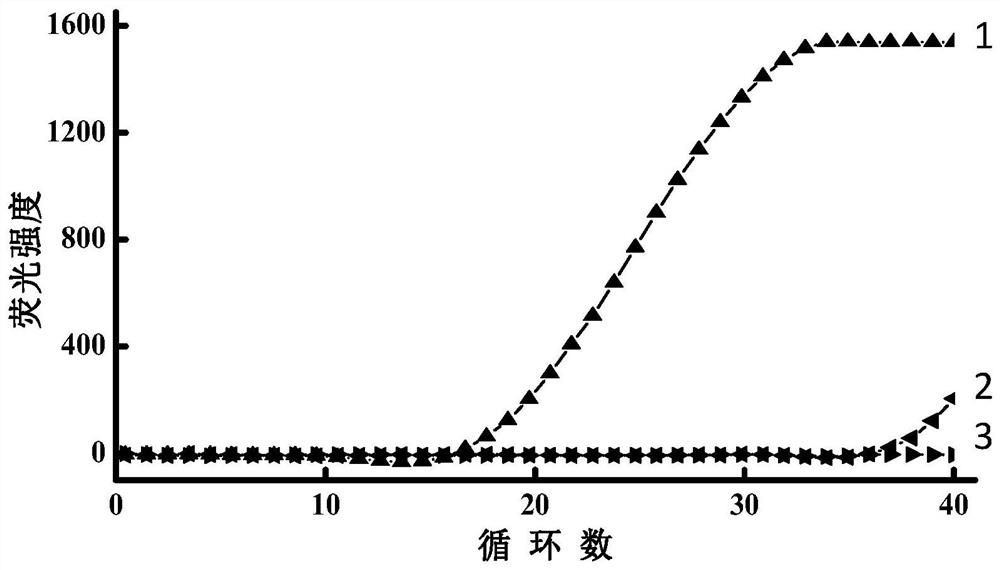
[0060] Example 2 Using linear probes to detect different concentrations of miRNA
[0061] This embodiment uses miRNA155 as the target (ie SEQ ID NO.1), using miRNA155 linear probe 1 (ie SEQ ID NO.2), PCR-F1 (ie SEQ ID NO.3), PCR-R1 (ie SEQ ID NO. NO.4) Perform reverse transcription and PCR amplification reactions, and detect different concentrations of target miRNA155 through fluorescent signals. miRNA122 (ie, SEQ ID NO.5) was used as a negative control, and water instead of the target was used as a blank control. Specific steps are as follows:
[0062] 1. Reverse transcription: i) prepare 15uL of reverse transcription reaction base solution; 7 , 3×10 6 , 3×10 5 , 3×10 4 , 3×10 3 , 3×10 2 , 3×10 1 , 3 copies of the target miRNA155, and mix the reaction solution; iii) place it on a PCR instrument, react at 42°C for 15min, and inactivate the reverse transcriptase activity at 85°C for 5min.
[0063] 2. Quantitative PCR amplification: the same as in Example 1.
[0064] F...
Embodiment 3
[0065] Embodiment 3 verifies the effect of linear probe d area and b area
[0066] The linear probe of the present invention includes three regions a, b and c, and may also include region d. The nucleotide sequence of the a region is complementary to the target nucleic acid, the nucleotide sequence of the b region assists loop formation by reducing steric hindrance, and the nucleotide sequence of the c region is identical to the 5' end of the target nucleic acid. The nucleotide sequence of the region d is a nucleotide sequence extending the length of the probe. By adjusting the GC content of d region and b region, the annealing temperature of upstream and downstream PCR primers is similar.
[0067] In this example, miRNA155 was used as the target miRNA (ie, SEQ ID NO.1), and corresponding linear probes and PCR primers were designed for reverse transcription and PCR amplification reactions, and the functions of the d and b regions of the probe were verified by fluorescent sign...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com