Application of EAF1 protein in regulation and control of anthocyanin synthesis and flowering time
A technology of flowering time and anthocyanin, applied in the fields of application, angiosperms/flowering plants, chemical instruments and methods, etc., can solve problems such as unclear molecular basis
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0095] Embodiment 1, the cloning of EAF1 gene
[0096] 1. Phenotype analysis of mutant eaf1
[0097] Take 30 mutant eaf1 seeds or Medicago truncatula R108 seeds, vernalize them at 4°C for 7 days, plant them in nutrient soil, and culture them in alternating light and dark (16h light / 8h dark), observe the phenotype and make statistics.
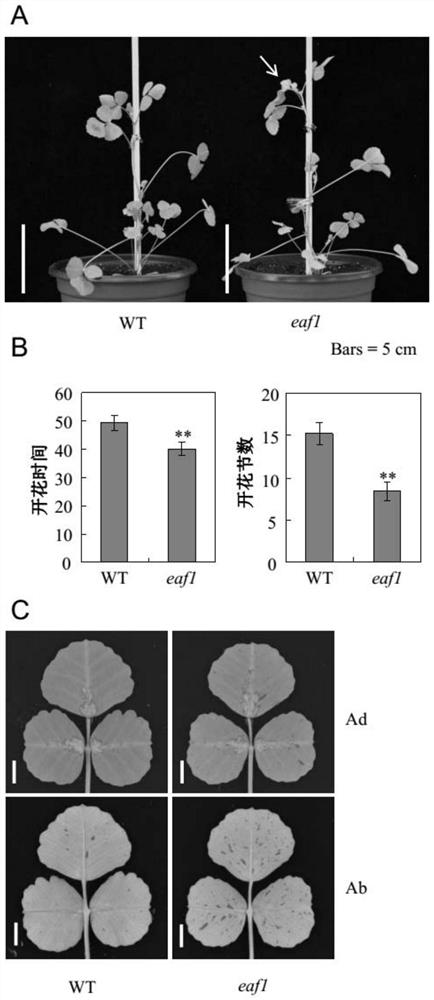
[0098] See the growth status of some mutants eaf1 and Medicago truncatula R108 when light and dark were alternately cultured for about 40 days figure 1Middle A (WT is Medicago truncatula R108, eaf1 is mutant eaf1). The results showed that the mutant eaf1 had flowered and had grown to 8 nodes on average after 40 days of alternating light and dark culture, while Medicago truncatula R108 had not flowered.
[0099] Count the flowering time and the number of nodes of each strain of Medicago truncatula (that is, the number of days of growth and the number of nodes of Medicago truncatula when the first flower fully opened), and then take the average...
Embodiment 2
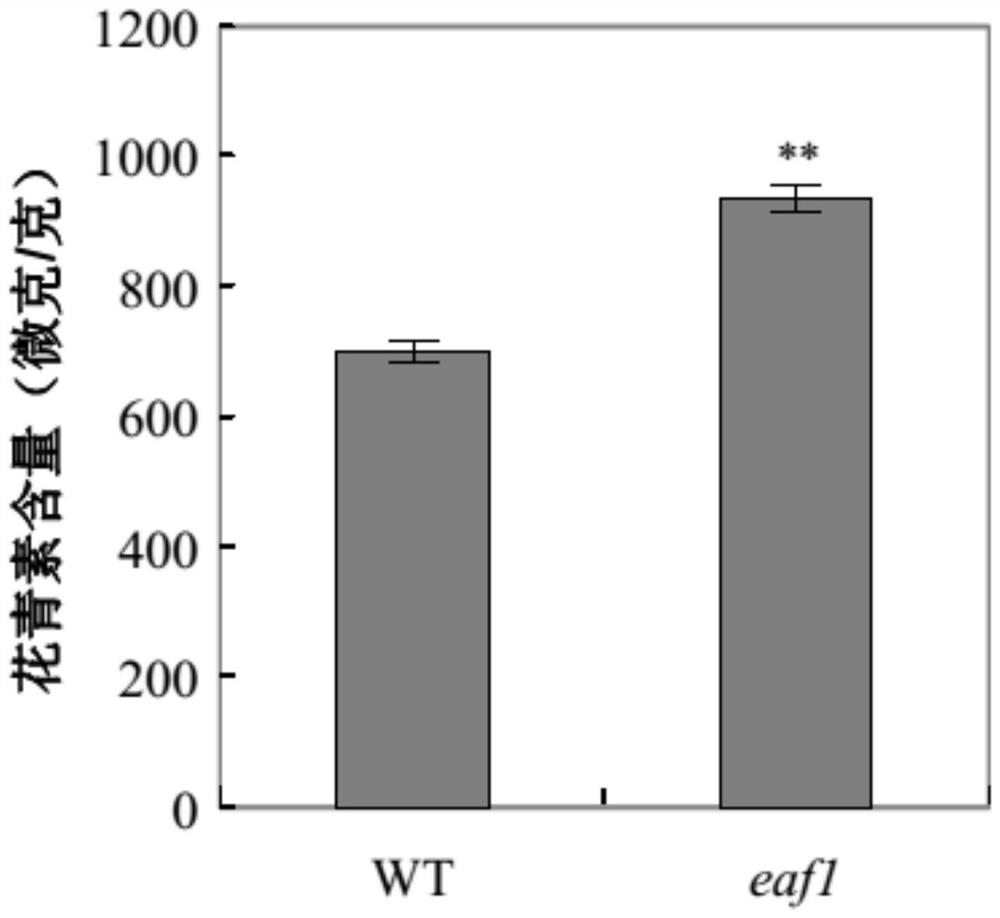
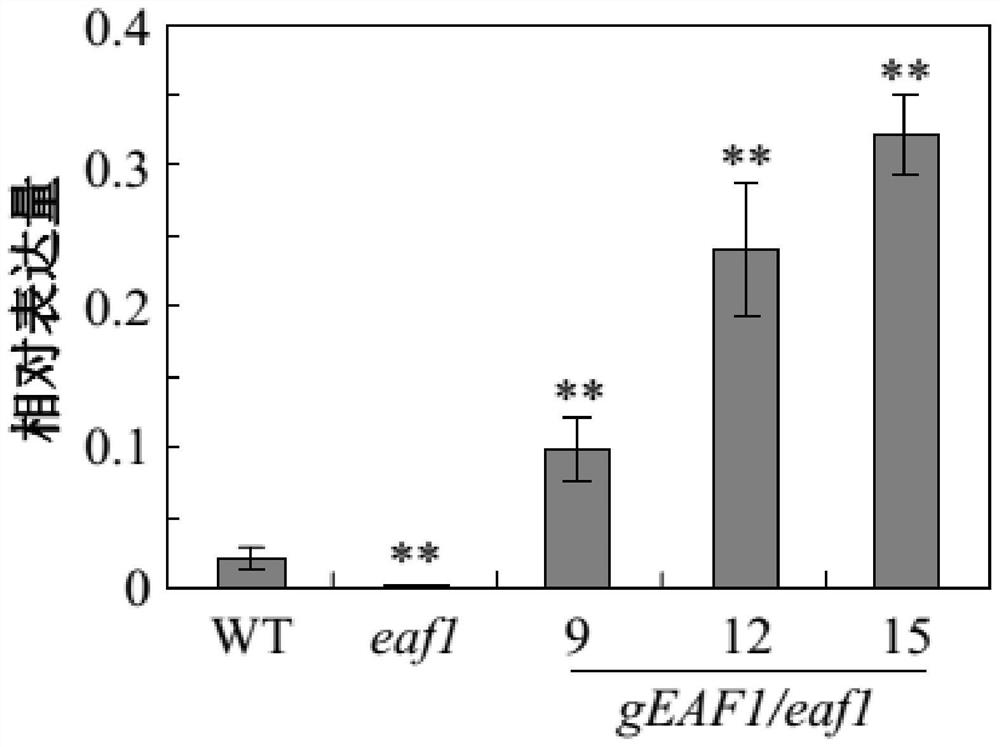
[0119] Embodiment 2, acquisition of complementary strains, phenotypic identification and detection of anthocyanin content
[0120] 1. Construction of recombinant plasmid pEAF1pro::EAF1 gDNA
[0121] 1. Using the genomic DNA of Medicago truncatula R108 as a template, using primer EAF1-EcoRI-F: 5′-CCATGATTAC GAATTC GAGTAAAGGCAAACACCACA-3' (the underline is the recognition site of restriction endonuclease EcoRI) and primer EAF1-KpnI-R: 5'-TCGACAGATCCCCG GGTACC A primer pair consisting of TGACGAACCTACCTTTACGT-3' (underlined is the recognition site of restriction endonuclease KpnI) was amplified to obtain a PCR amplification product of about 3250 bp.
[0122] 2. Digest the PCR amplified product obtained in step 1 with restriction endonucleases EcoRI and KpnI, and recover the digested product.
[0123] 3. The pCAMBIA2300 vector was digested with restriction enzymes EcoRI and KpnI, and the vector backbone of about 9850 bp was recovered.
[0124] 4. Ligate the digested product wi...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com