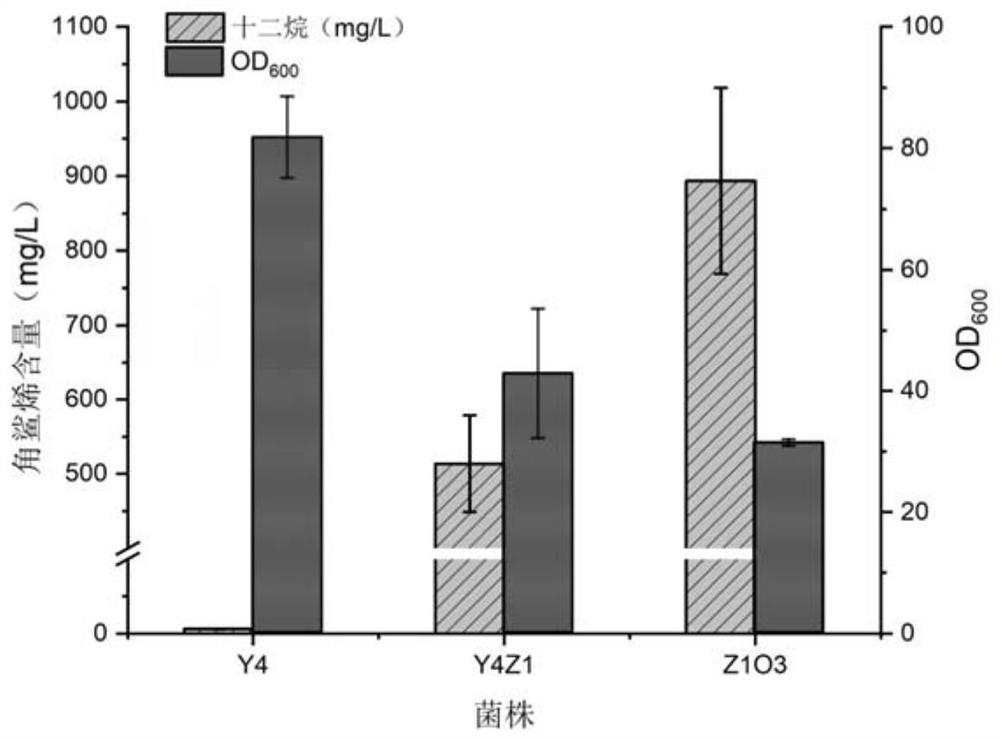
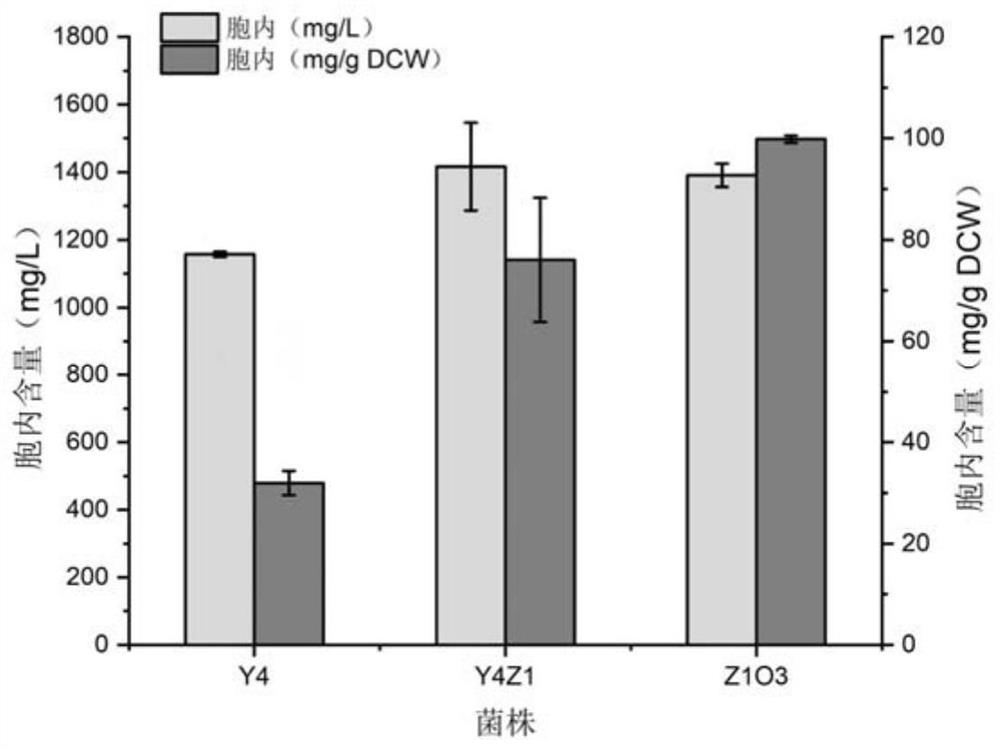
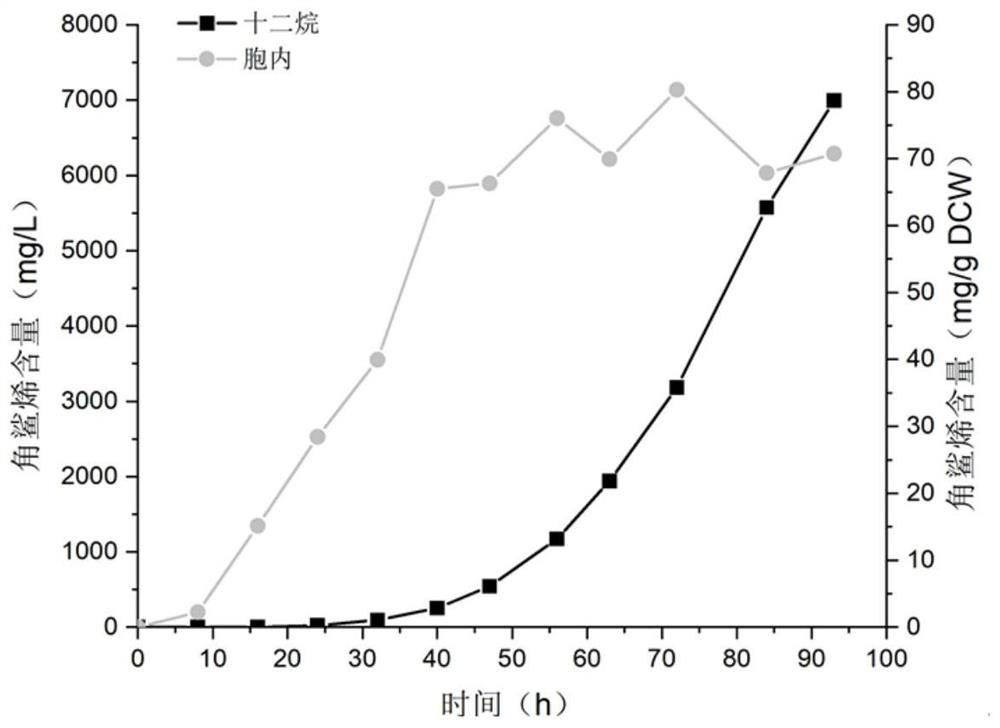
Construction and application of saccharomyces cerevisiae strain for extracellular transport of vitamin D3 precursor squalene
A technology of Saccharomyces cerevisiae and recombinant Saccharomyces cerevisiae, which is applied in the field of Saccharomyces cerevisiae strain construction to achieve enhanced effects, broad application prospects, and strong production performance
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0055] Example 1: Construction of Saccharomyces cerevisiae strain Y1
[0056] Specific steps are as follows:
[0057] (1) Artificially synthesized gene fragment P GPD -tHMG1-T ADH1 (The nucleotide sequence is shown in SEQ ID NO.1);
[0058] Using the Saccharomyces cerevisiae BY4741 genome as a template, the primer sequences described in Table 1 were used to amplify the gene fragment ROX1-UP with primers ROX1-UP-F and ROX1-UP-R;
[0059] The gene fragment ROX1-DOWN was amplified with primers ROX1-DOWN-F and ROX1-DOWN-R;
[0060] Using the plasmid pMHyLp-His (nucleotide sequence shown in SEQ ID NO.2) as a template, the ROX1-His fragment was amplified using primers ROX1-loxH-F and ROX1-loxH-R.
[0061] (2) The four fragments P in step (1) GPD -tHMG1-T ADH1 , ROX1-UP, ROX1-DOWN, and ROX1-His were fusion-PCRed by PCR, and the correct band obtained by running the gel was recovered by cutting the gel to obtain the fusion gene fragment ΔROX1-P GPD -tHMG1-T ADH1 .
[0062] (3)...
Embodiment 2
[0064] Example 2: Construction of Saccharomyces cerevisiae strain Y2
[0065] (1) Artificially synthesized gene fragment P TEF1 -IDI-T CYC1 (The nucleotide sequence is shown in SEQ ID NO.4);
[0066] Using the Saccharomyces cerevisiae BY4741 genome as a template, the primer sequences shown in Table 1 were used to amplify the gene fragment 911b-UP with primers 911b-UP-F and 911b-UP-R;
[0067] Amplify the gene fragment 911b-DOWN with primers 911b-DOWN-F and 911b-DOWN-R;
[0068] Using the plasmid pMHyLp-His as a template, the INO2-His fragment was amplified with primers IDI-loxH-F and IDI-loxH-R.
[0069] (2) The four fragments P in step (1) TEF1 -IDI-T CYC1 , 911b-UP, 911b-DOWN, IDI-His use PCR for fusion PCR, and the correct band obtained by running the gel is recovered by cutting the gel to obtain the fusion gene fragment 911b-P TEF1 -IDI-T CYC1 .
[0070](3) Transform the gene fragment in step (2) into the competent Y1 strain prepared in Example 1, culture it on the...
Embodiment 3
[0072] Example 3: Construction of Saccharomyces cerevisiae strain Y3
[0073] Specific steps are as follows:
[0074] (1) Artificially synthesized gene fragment P GPD -tHMG1-T ADH1 -P TEF1 -ERG20-T CYC1 (The nucleotide sequence is shown in SEQ ID NO.5);
[0075] Using the Saccharomyces cerevisiae BY4741 genome as a template, the primer sequences described in Table 1 were used to amplify the gene fragment ERG20-UP with primers ERG20-UP-F and ERG20-UP-R;
[0076] The gene fragment ERG20-DOWN was amplified with primers ERG20-DOWN-F and ERG20-DOWN-R;
[0077] Using the plasmid pMHyLp-His as a template, the ERG20-His fragment was amplified with primers ERG20-loxH-F and ERG20-loxH-R.
[0078] (2) The four fragments P in step (1) GPD -tHMG1-T ADH1 -P TEF1 -ERG20-T CYC1 -P PGK , ERG20-UP, ERG20-DOWN, and ERG20-His use PCR for fusion PCR, and the correct bands obtained by running the gel are recovered by cutting the gel to obtain the fusion gene fragment P GPD -tHMG1-T ADH...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com