Biomarkers for detecting colorectal cancer or adenoma and methods thereof
A technology for rectal adenoma and colorectal cancer, applied in the field of biomarker detection of colorectal cancer or adenoma, can solve problems such as low accuracy
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0175] Example 1 Semi-quantitative untargeted metabolomic analysis in serum from the discovery cohort reveals significantly altered metabolites in CRC and adenoma patients
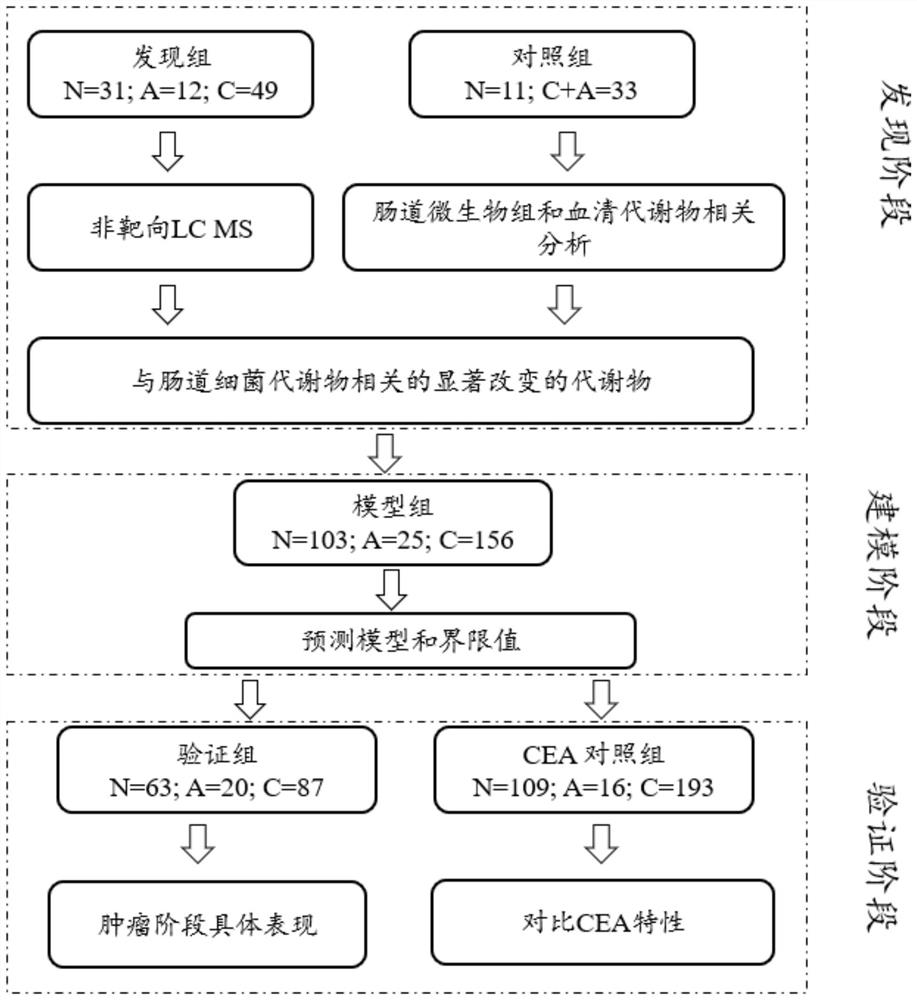
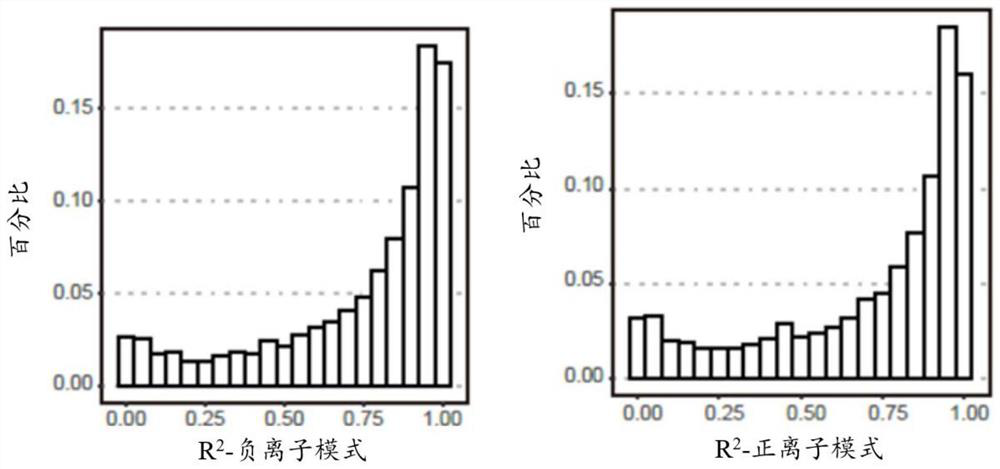
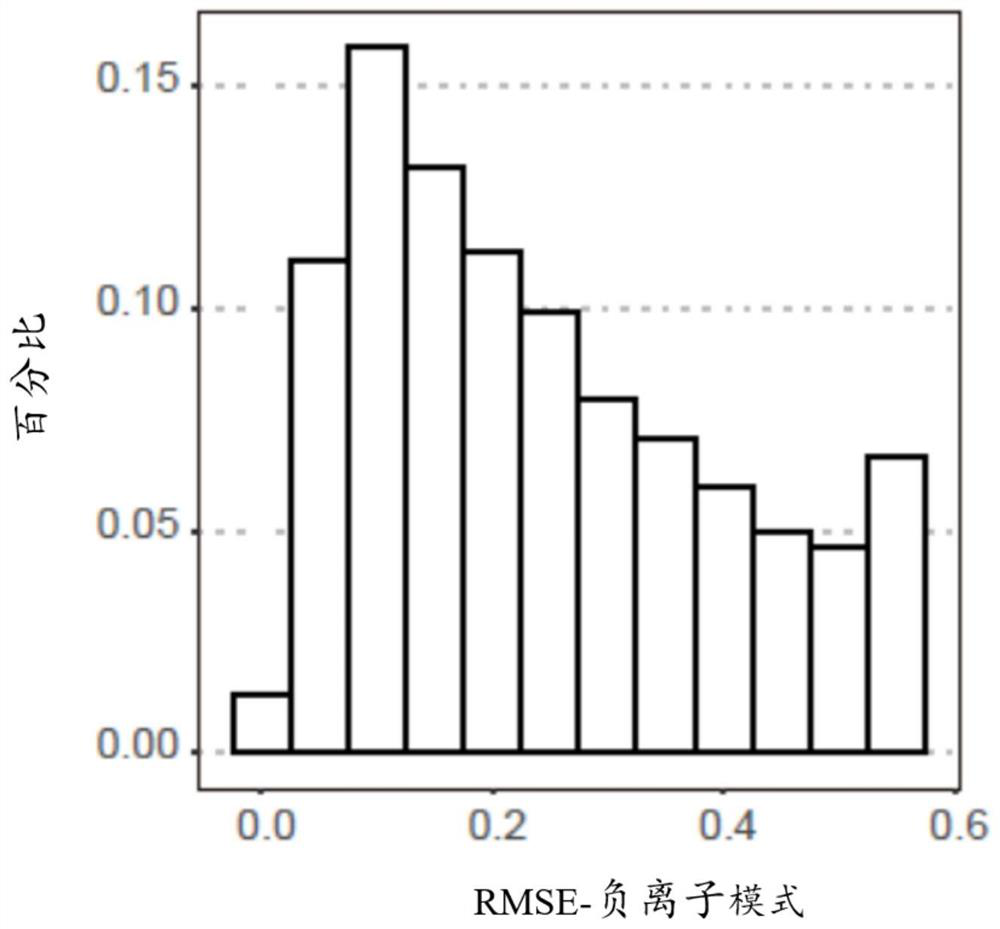
[0176] The relationship between the serum metabolome and colorectal adenoma or colorectal cancer was determined by untargeted metabolome profiling of the discovery panel by LCMS. Figure 1A is a schematic diagram showing an overview of the experimental design and analysis procedures according to some embodiments of the present application. Such as Figure 1A As shown, the discovery group was divided into normal group, adenoma group and colorectal cancer group. Low-abundance signals (eg, mean abundance Figure 1B is an analysis plot of the distribution of R2 values for non-targeted LC-MS features in negative ion mode and positive ion mode. The R2 values of the linear regression model between the expected and measured mixing ratios for each metabolite detected in the negative and positive ion modes are...
Embodiment 2
[0180] Example 2 Determination of significantly altered gut microbiome-associated serum metabolites in patients with colorectal abnormalities
[0181] Figure 2A is the analysis plot of the program integrating the analysis of fecal metagenome and serum metabolome in the matched group. In total, data from 44 subjects (11 normal and 33 with colorectal abnormalities) passed quality control. Taxonomic analysis of metagenomic data revealed 12,455 microbiome species. Figure 2B is a bar graph of the 15 OUTs in each individual at the species level. Among the top 15 most abundant species, elevated enterotoxigenic Bacteroides fragilis (ETBF), considered a key pathogen for CRC initiation, was observed. Such as Figure 2C is a plot of the relative abundance of several CRC-associated gut microbiome species in matched cohorts of patients with normal and colorectal abnormalities. Such as Figure 2C As shown, the abundance of several other CRC-promoting species, including Fusobacterium...
Embodiment 3
[0183] Example 3 Prediction of colorectal abnormalities in the discovery cohort based on a list of gut microbiome-associated serum metabolites
[0184] Based on these gut microbiome-associated serum metabolites above, the LASSO algorithm was performed to find key metabolite biomarkers of colorectal abnormalities. The LASSO algorithm with 10-fold cross-validation (CV) was used for feature selection from previously determined serum metabolomic data and gut microbiome metabolomic data. 322 metabolite signatures were significantly altered between normal and CRC or adenoma samples (adjusted p < 5E-3) and were significantly associated with the gut microbiome (p < 1E-3, FDR ≤ 18%). Using group voting, more than 75% of the 200 LASSO runs involved 32 metabolite signatures. Their chemical structure annotations, including MS2 transitions if identifiable, were established by MS / MS spectrum matching as previously described. As mentioned above, Table 1 lists 8 metabolite signatures out of...
PUM
| Property | Measurement | Unit |
|---|---|---|
| Sensitivity | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com