SNP locus combination for detecting tomato verticillium wilt resistance and application thereof
A Verticillium wilt and disease resistance technology, applied in the field of plant biology, can solve the problems of linkage redundancy, insufficient versatility of SNP sites, invalid selection of target genes, etc., achieve accurate detection, reduce manual inoculation identification and field transplanting work Quantity, the effect of improving breeding efficiency
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0074] The screening of embodiment 1 SNP site combination
[0075] 1. Experimental materials
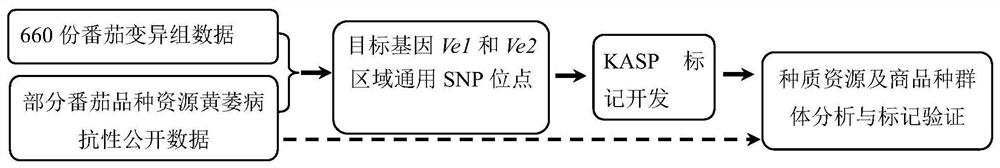
[0076] The variant group data of 660 representative tomato germplasm resources from different sources worldwide were selected for SNP site screening in this embodiment. The source of the tomato variant group data is mainly based on the previous work of the project team where the inventor belongs. (Lin,T.,Zhu,G.,Zhang,J.et al.Genomic analyzes provide insights into the history of tomato breeding[J].Nat Genet,2014,46:1220~1226; Tieman D,Zhu G,Resende M F R , et al. A chemical genetic roadmap to improved tomato flavor [J]. Science, 2017, 355(6323): 391). Among them, the genotype data of some tomato germplasm resources and the resistance phenotype data of Ve1 and Ve2 genes come from the existing public database (https: / / solgenomics.net / ).
[0077] 2. Screening of SNP site combinations
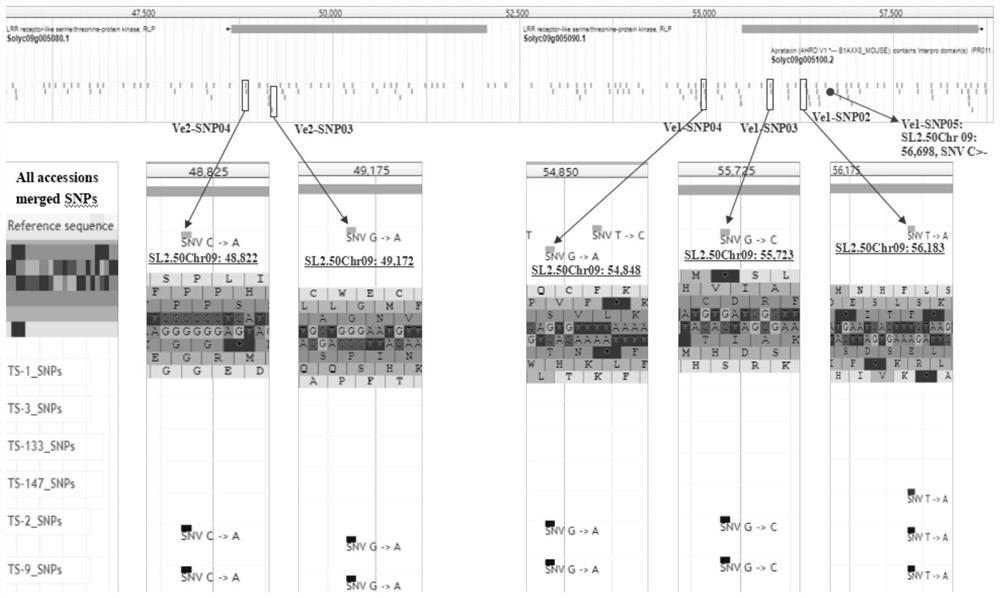
[0078] Use target genes Ve1 (Solyc09g005090, database source: https: / / solgenomics.net / locus / 1531 / vie...
Embodiment 2
[0082] Example 2 Primer Synthesis and Kit Preparation
[0083] 1. Primer design and screening
[0084] According to the flanking sequences of the Ve1-SNP01 site to Ve1-SNP04 site and the Ve2-SNP01 site to Ve2-SNP05 site provided in Example 1, for each SNP site, the principle of design and development according to the KASP marker (target product 80-150bp, the primers specifically match the target region on the reference genome and are located in non-SNP dense regions, avoiding complex sequence regions with high A\T or G\C content, etc.), use Primer3.0 software to design two upstream SNP sites A forward primer and a reverse primer were designed downstream.
[0085] Tomato samples to be tested include some germplasm resources of known genotypes and Verticillium wilt resistance phenotypes and 21 samples randomly selected from breeding materials as resistant, susceptible or heterozygous controls, and finally added 3 ddH 2 O is used as the NTC blank control, a total of 24 copies. ...
Embodiment 3
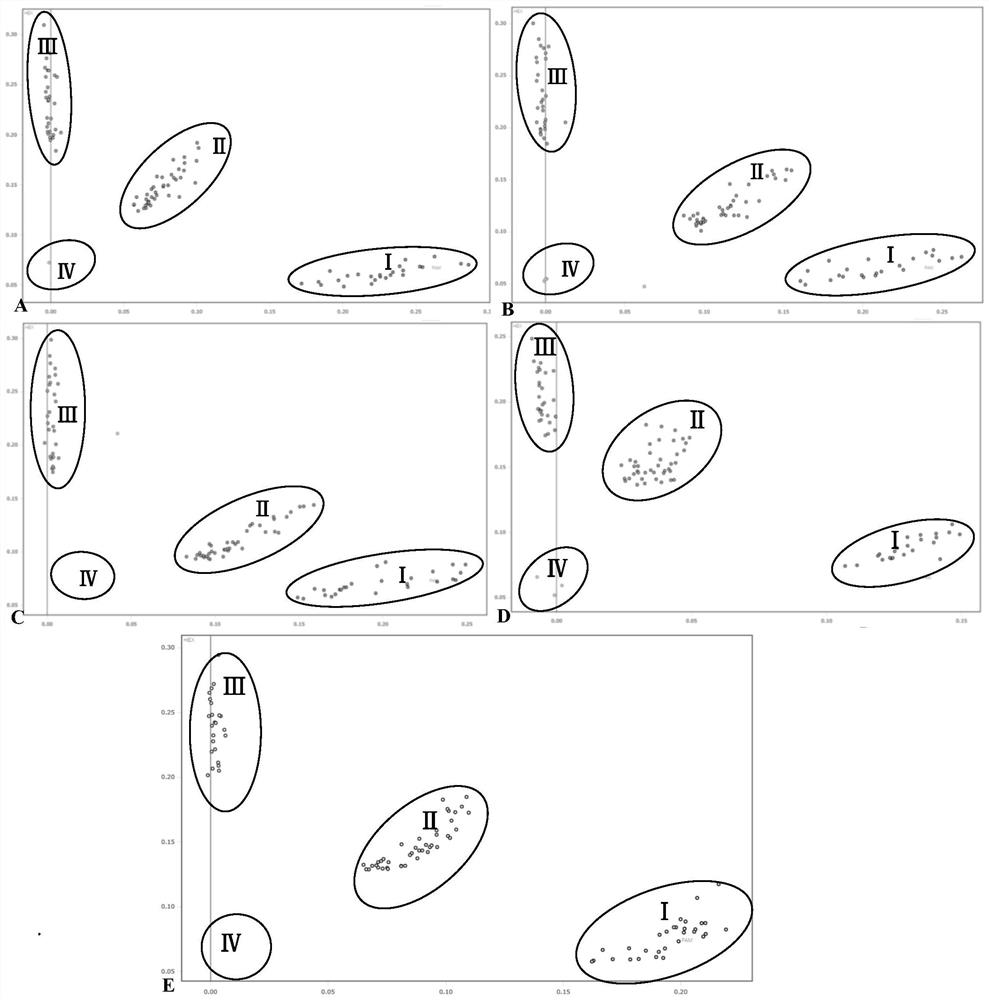
[0098] Example 3 Verification and Breeding Application of Tomato Verticillium Wilt Resistance Genes Ve1 and Ve2 Highly Efficient KASP Markers
[0099]In this example, based on the SNP loci provided in Example 1, the kit (including primer combinations) provided in Example 2 was used to carry out the verification and breeding application of tomato Verticillium wilt resistance genes Ve1 and Ve2 high-efficiency KASP markers.
[0100] Select 381 representative tomato samples including mainstream commercial varieties, core public germplasm of the National Resource Bank, breeding intermediate materials, new hybrid combinations, etc., involving large fruit tomatoes, cherry tomatoes, cluster tomatoes, fresh tomatoes, processed tomatoes and Various types of cultivated tomatoes such as farm species (local varieties). Among them, 177 have obtained Ve1 and Ve2 genotype data through third-party commercial institutions using closely linked SCAR markers (including 15 tomato germplasm resource...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com