Recombinant saccharomyces cerevisiae strain capable of simultaneously expressing IFNa14 protein and human hepatitis B virus S protein as well as preparation method and application
A technology for recombining Saccharomyces cerevisiae and hepatitis B virus is applied in the field of preparation of recombinant Saccharomyces cerevisiae strains, which can solve the problems of limited treatment time and low curative effect, and achieve the effects of low cost, large-scale production, and good application and development prospects.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0035] Example 1. Construction of GPD-HBVs-TU and GPD-IFNα14-TU vectors
[0036] (1) Amplification of HBVs and IFNα14 genes
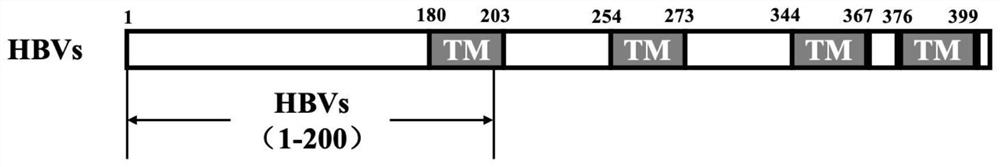
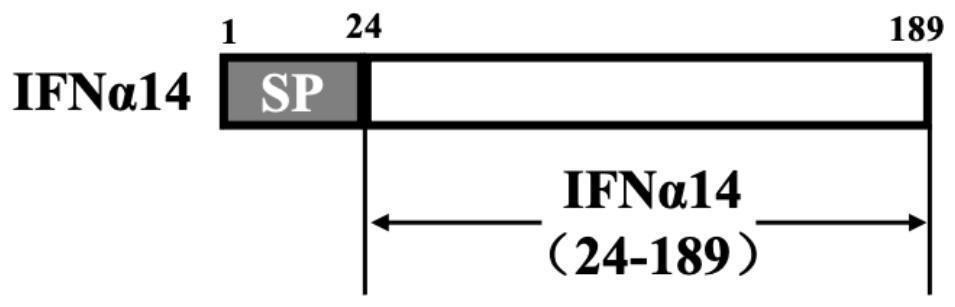
[0037] According to the sequence structure of HBVs and IFNα14 gene ( figure 1 , 2 shown), design and synthesize primers, TU-HBVs-F (SEQ ID NO.7: gacgataagg taccaggatc catgggagga tggtcatcaa agc) and TU-HBVs-R (SEQ ID NO.8: tgctggatat ctactggatc cgagtatacg tgtcaggagg aagaatc), TU-IFNα14-F (SEQ ID NO. 9: gatgtgcttc gattccccgg gtgtaacctt tcacaaacac attcact) and TU-IFNα14-R (SEQ ID NO. 10: atctgtaagt ctagacccgg ggtcctttct tctcagccgc tt). According to the reported HBVs gene sequence (Genbank number AF384371.1) and IFNα14 gene sequence (Genbank number NM_002172.3), the genes of HBVs protein and IFNα14 protein were optimized and artificially synthesized, and pET28a(+)-HBVs plasmid was constructed and pET28a(+)-IFNα14, using the plasmid as a template to amplify the coding genes of HBVs (1-200) and IFNα14 (24-189). The PCR amplification system is:
[0038] ...
Embodiment 2
[0045] Example 2. Construction and detection of recombinant Saccharomyces cerevisiae strains expressing IFNa14 protein and human hepatitis B virus S protein simultaneously
[0046] (1) Construction of Yeast Transformation Fragment
[0047] The vector GPD-HBVs-TU was cut with BsaI, and at the same time, the homology arm plasmids (URR1 and URR2) and the selectable marker plasmid (LEU) were cut with BsmB I. Refer to the experimental steps of Dai's research group [6] , URRs homology arms, LEU selective tags, and GPD-HBVs-TU transcription units were spliced according to the specific prefix and suffix sequences, T4 ligase was ligated overnight at 16°C, and the spliced products were used for yeast transformation.
[0048] The vector GPD-IFNα14-TU was cut with BsaI, and at the same time, the homology arm plasmids (SUR1 and SUR2) and the selectable marker plasmid (Trp) were cut with BsmB I. Refer to the experimental steps of Dai's research group [6] , the SURs homology arm, Trp ...
PUM
| Property | Measurement | Unit |
|---|---|---|
| molecular weight | aaaaa | aaaaa |
| molecular weight | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com