mRNA-based vaccine against coronavirus and preparation method thereof
A gene and N-terminal technology, applied in the field of vaccines against coronavirus and its preparation, can solve the problems of inability to effectively take into account virus gene mutations, low proportion of antigenic determinants, and large molecular weight of expressed proteins, so as to reduce cytotoxicity and reduce Vaccine toxicity, effect of guaranteeing immune effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
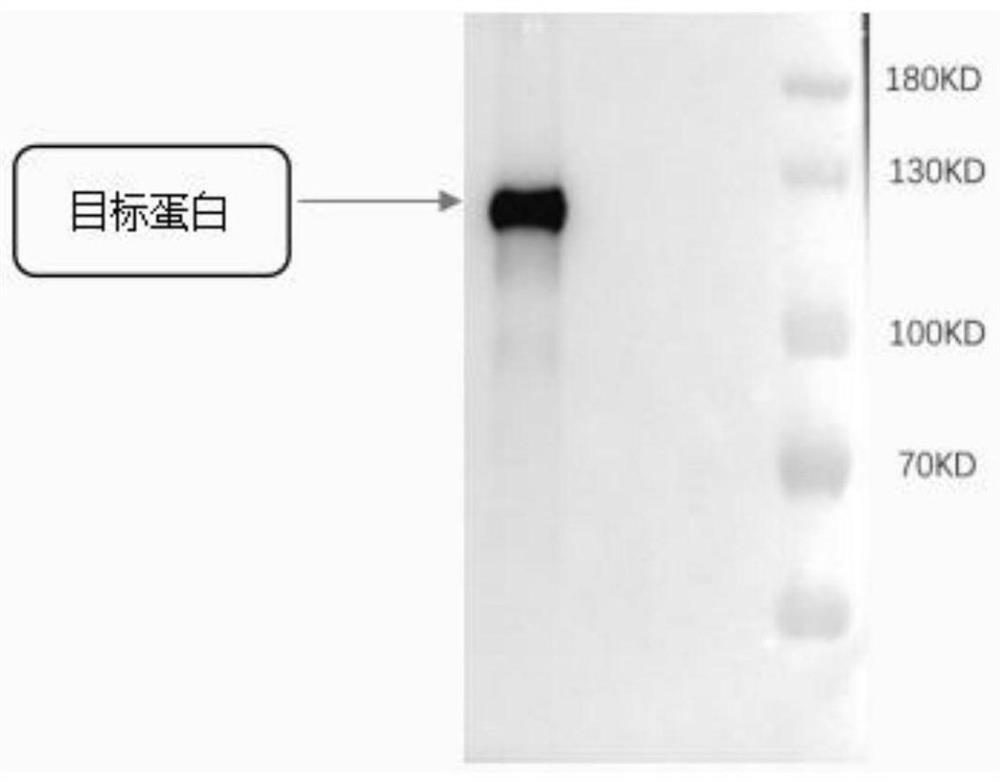
Image
Examples
preparation example Construction
[0060] The preparation method for coronavirus vaccine based on mRNA comprises the steps:
[0061] The step of preparing mRNA-RBD+NTD+RBD preparation comprises:
[0062] Step a, synthesizing RBD+NTD+RBD gene fragments:
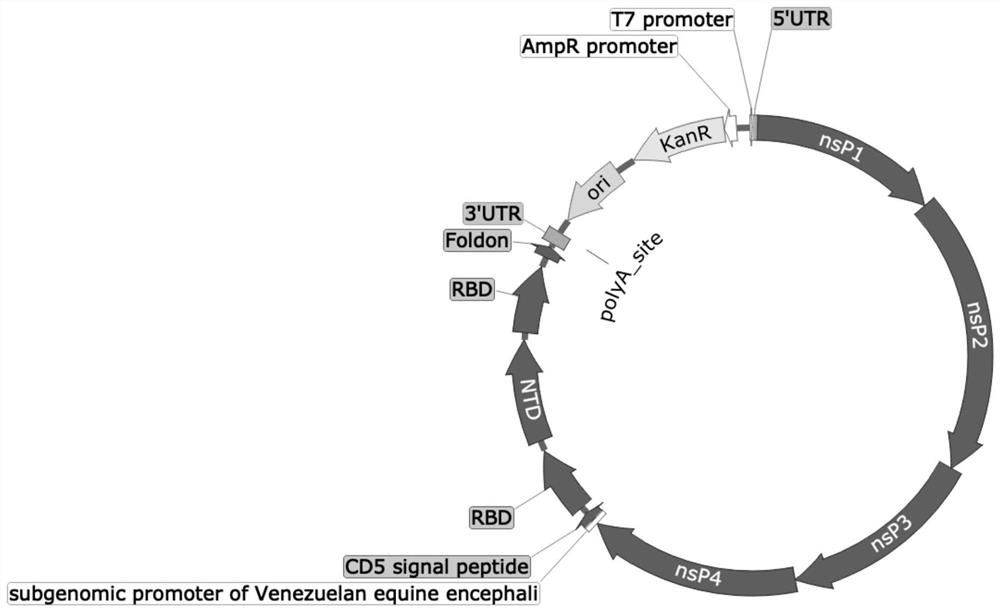
[0063] Query the amino acid sequence of Spike and then design the secretion signal peptide, receptor binding domain (RBD) + N-terminal domain (NTD) + receptor binding domain (RBD) + Foldon gene sequence, and optimize the codon according to the human preferred codon table For the optimized sequence, a promoter and an ApaI restriction site GGGCCC are added upstream of the optimized sequence, and a NotI restriction site GCGGCCGC is added downstream, and the designed sequence is obtained by synthesis; the gene sequence is SEQ ID No. in the sequence list. 2 shown;
[0064] Step b, construct TC-83 self-replicating vector:
[0065] Design self-replicating mRNA sequence according to the genome of alpha virus family, described self-replicating mRNA sequence comprises...
Embodiment 1
[0087] Embodiment 1 RBD+NTD+RBD gene fragment synthesis
[0088] Query Spike (SARS-CoV-2) and British variant B.1.1.7, South African variant B.1.351, Brazilian variant P.1 and Indian variant B.1.617 protein genes from NCBI, and then design secretion signal peptide, receptor binding region (RBD)+N-terminal region (NTD)+receptor binding region (RBD)+Foldon sequence gene, and then optimize according to human codons, and add ApaI restriction site and promoter upstream, and add NotⅠ downstream Restriction sites, and finally obtain the DNA sequence directly by synthesis (obtained in the form of cloning plasmid pUC57-RBD+NTD+RBD provided by the company), (the sequence is shown in SEQ ID No.2). The encoded amino acid sequence is shown in SEQ ID No.1.
[0089]
Embodiment 2
[0090] Example 2 Construction of TC-83 self-replicating vector
[0091] The self-replicating mRNA is designed based on the genome of the Venezuelan Equine Encephalomyelitis Virus (TC83 Venezuelan Equine Encephalitis Virus, VEEV) in the alphavirus family, which contains genes that encode the self-replicating components of the alphavirus, but lacks the coding for the production of infectious The structural protein of the alpha virus particle, and the constructed plasmid is used as a template to amplify, and the designed sequence is directly obtained by synthesis (the sequence is shown in SEQIDNo.3).
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com