Method for constructing HCMV encoding miRNA deletion strain based on Crispr-cas9 technology and application thereof
A gene and target technology, applied in the field of constructing HCMV-encoded miRNA deletion strains, can solve the problems of inability to miRNA regulation, long time period, and high equipment requirements
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
preparation example Construction
[0050] The preparation method of the LB plate with ampicillin resistance is: 1) get ampicillin (0.5g / bottle) (Yuekang Pharmaceutical Group Co., Ltd. product, batch number: 22081002), add 5mL deionized water after autoclaving, directly Mix well to obtain a stock solution of ampicillin with a concentration of 100 mg / mL, and freeze it at -20°C for later use; 2) Accurately weigh 10 g of tryptone, 5 g of yeast extract, 10 g of NaCl, and 15 g of agar powder, add deionized water to make up to 1 L, and autoclave Bacteria; after cooling to 50°C, add ampicillin stock solution at a ratio of 1:1000, mix well, quickly pour it into a high-pressure plate and let it stand, after cooling and solidifying, invert the plate to obtain an LB plate with ampicillin resistance, 4° Refrigerate and set aside.
[0051] The preparation method of the 10% fetal calf serum DMEM culture medium is: in the DMEM medium, add serum according to 10% volume under sterile conditions, the serum product number (04-001-...
Embodiment 1
[0057] The preparation of embodiment 1 △ miRNA-HCMV virus strain
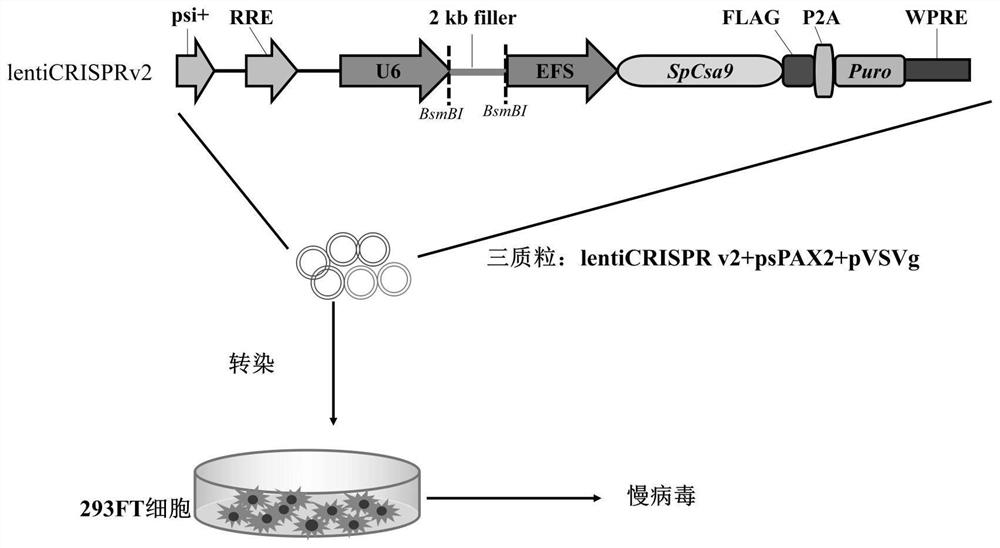
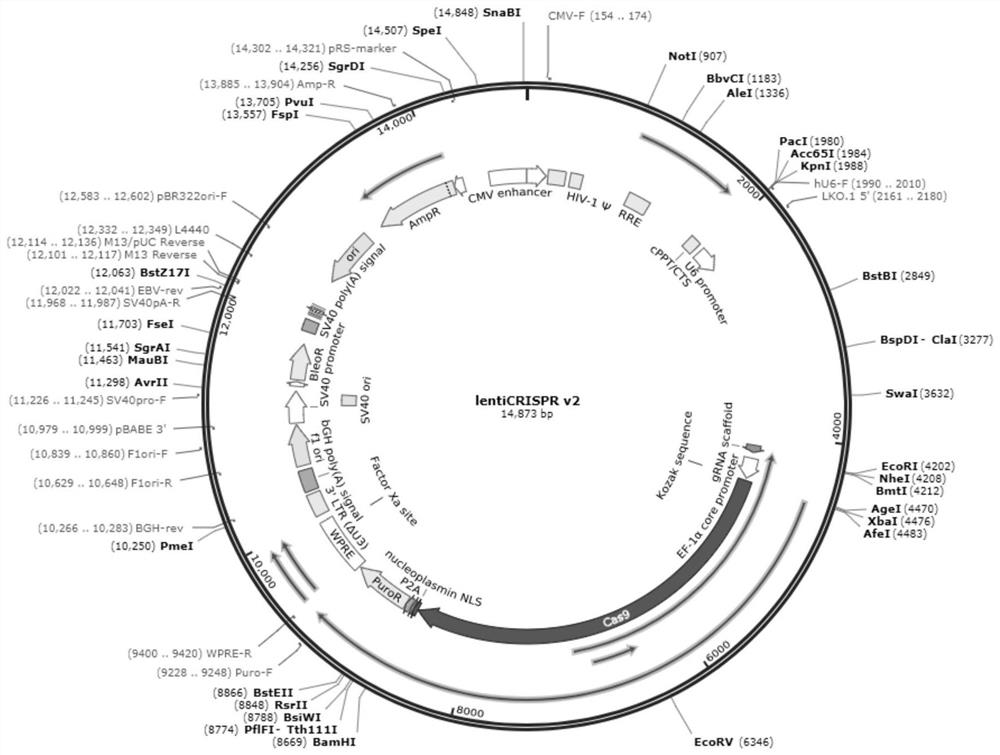
[0058] For the preparation flow chart of expressing sgRNA-Cas9 lentivirus, see figure 1 ,Specific steps are as follows:
[0059] 1. Construction of vector pLenti-US33as-5p-sgRNA
[0060] 1. Design the target sequence of miR-US33as-5p
[0061] The sequence of hcmv-miR-US33as-5p is as follows:
[0062] 5'-GGAUGUGCUCGGACCGUGACGGUG-3' (as sequence 1 in the sequence listing)
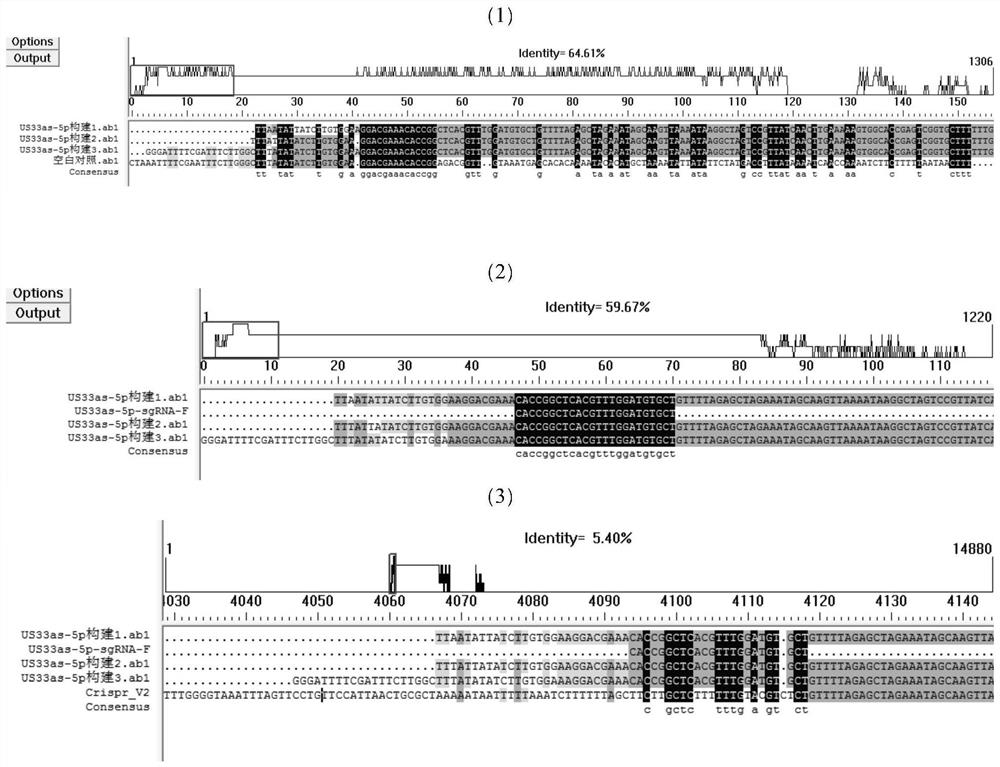
[0063] In order to knock out the hcmv-miR-US33as-5p gene, select sgRNA, its name is US33as-5p-sgRNA, and its nucleotide sequence is 5'-GCUCACGUUUGGAUGUGCU-3'. The coding strand sequence of the US33as-5p-sgRNA gene is: 5'-GCTCACGTTTGGATGTGCT-3' (such as the 1st-19th position of sequence 2 in the sequence listing)
[0064] For the US33as-5p-sgRNA gene, DNA primers F and R (underlined nucleotides are reverse complementary) were designed and synthesized by Beijing Aoke Biological Company.
[0065] F: 5'-CACCG GCTCACGTTTGGATGTGCT -3';
[0...
Embodiment 2
[0105] Example 2, hcmv-miR-US33as-5p target identification and verification of its protective effect on interferon
[0106] 1. Screening and identification of hcmv-miR-US33as-5p target
[0107] 1. hcmv-miR-US33as-5p target screening
[0108] Use prediction software to predict and analyze the target of hcmv-miR-US33as-5p (also known as hcmv-US33as-5p), the results are shown in Image 6 , and the predicted targets are the 13 genes in Table 1, respectively.
[0109] Table 1 The sequences of different primer pairs designed for the predicted target of hcmv-miR-US33as-5p
[0110]
[0111]
[0112] The targets predicted by the software were respectively constructed with dual luciferase reporter plasmids containing the predicted target 3'-UTR, and co-transfected with the hcmv-miR-US33as-5p mimic into 293FT cells, and the predicted target 3'-UTR was transferred and expressed 13 kinds of luciferase recombinant 293FT cells, 293FT cells transfected with predicted target 3'-UTR+NC-R...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com