Endonuclease and application thereof
A technology of endonuclease and amino acid, applied in the direction of hydrolase, introduction of foreign genetic material by using carrier, recombinant DNA technology, etc., to achieve the effect of broad application prospects
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment approach
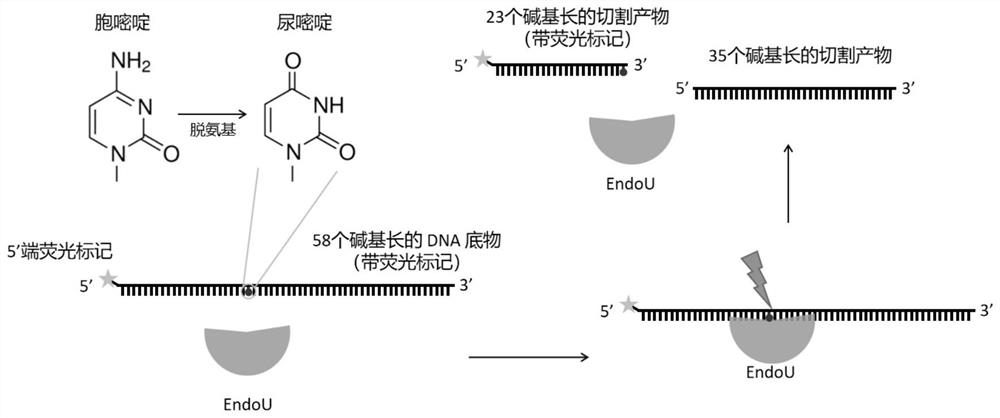
[0045] In one embodiment, the step of using the endonuclease to recognize deoxyuracil in the DNA containing deoxyuracil and cleave the first phosphodiester bond at the 3' end of the deoxyuracil comprises:
[0046] Provide DNA and endonuclease containing deoxyuracil;
[0047] The endonuclease, the DNA containing deoxyuracil, Tris hydrochloride, sodium chloride, dithiothreitol and glycerin are mixed and incubated, and the endonuclease is effective against the The deoxyuracil in the deoxyuracil-containing DNA recognizes and cleaves the first phosphodiester bond at the 3' end of the deoxyuracil.
[0048] The present invention will be further described below by specific examples.
Embodiment 1
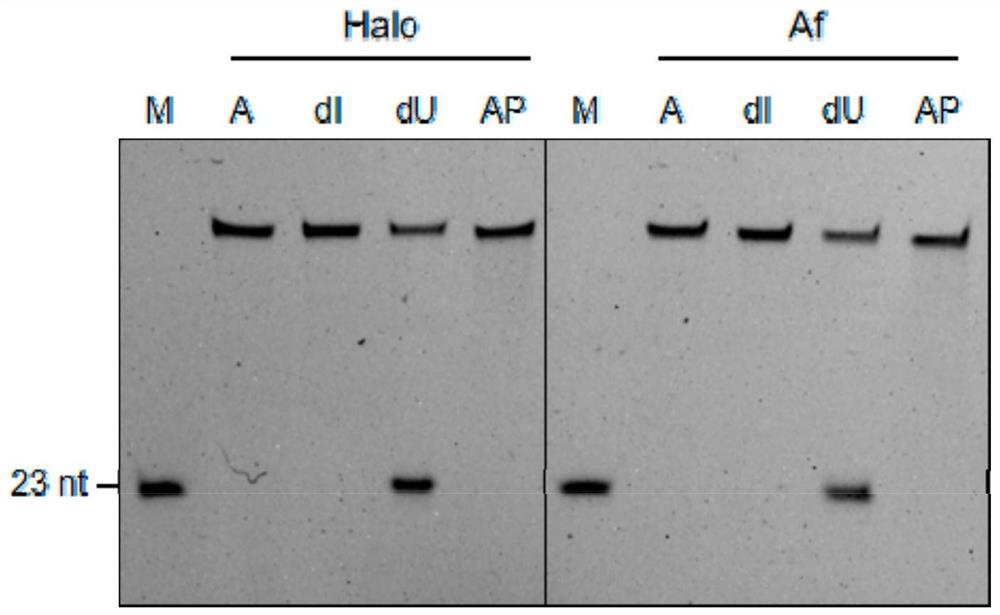
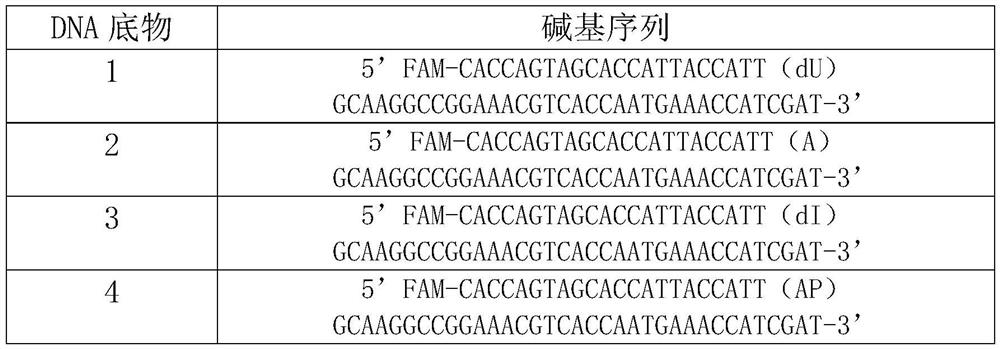
[0050] The base sequence of the single-stranded DNA substrate in the present embodiment is shown in Table 1 below:
[0051] Table 1. Base sequences of single-stranded DNA substrates
[0052]
[0053] (1) Synthesis of the endonuclease (the amino acid sequence of which is shown in SEQ ID NO: 1 and 2): the DNA encoding the endonuclease is synthesized through the artificial gene synthesis of the commercial company Beijing Liuhe Huada Gene Technology Co., Ltd. The sequence is inserted into an expression plasmid, and then heterologously expressed in Escherichia coli, and purified by nickel ion affinity chromatography to obtain a pure protein (the endonuclease) for use.
[0054] (2) Synthetic single-stranded DNA substrate:
[0055] Synthesize 58 bases, the 23rd position is deoxyuridine (dU), thymidine (A), deoxyinosine (dI), apurine pyrimidine (AP), and the 5' end has FAM fluorescently labeled single-stranded DNA substrates 1, 2, 3, 4. The base sequences of single-stranded DNA ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com