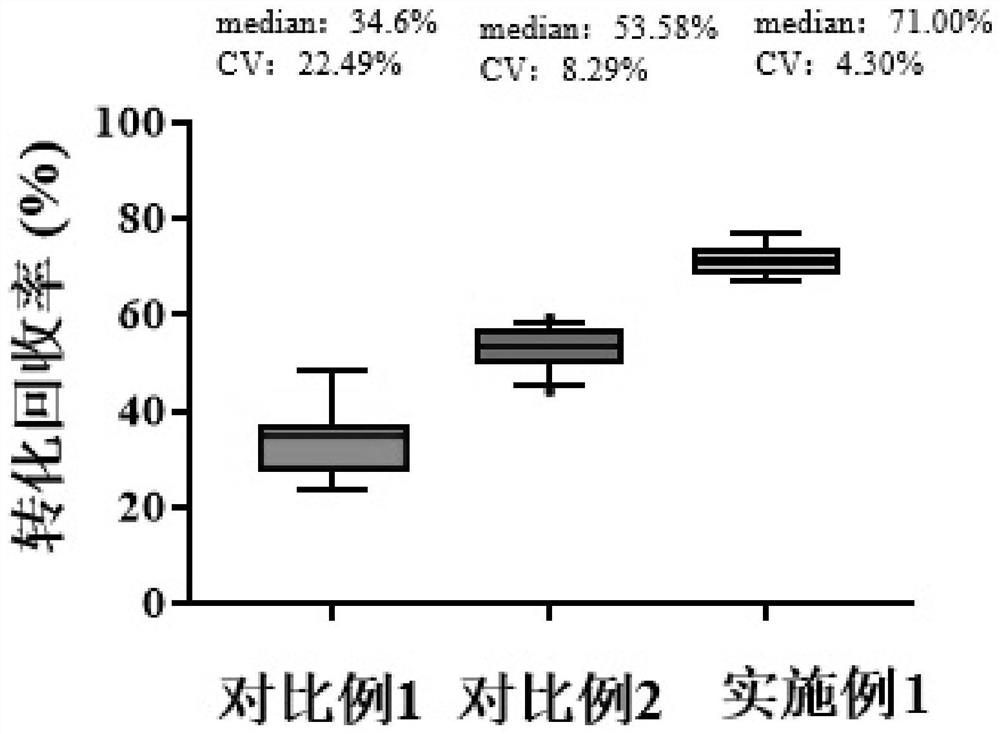
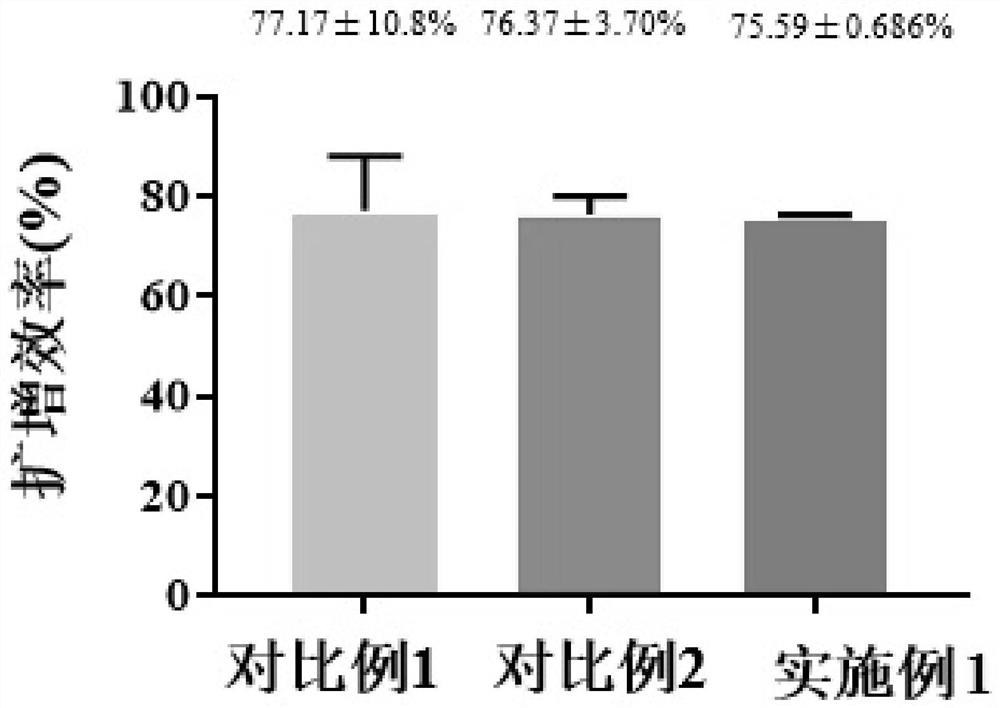
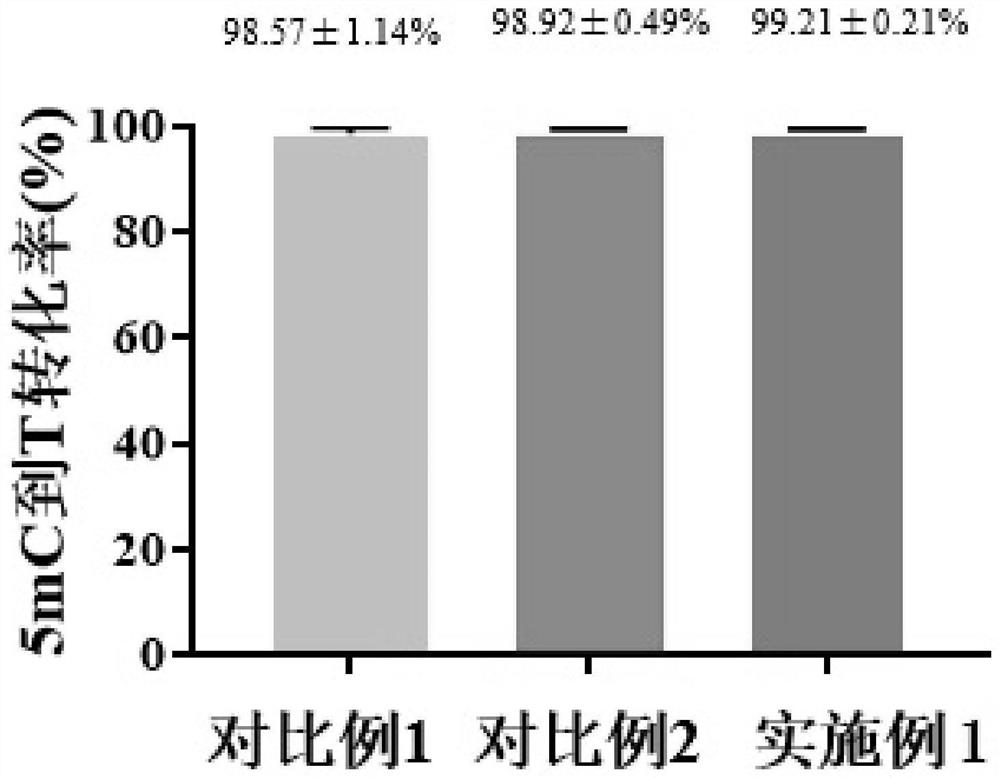
Method for reducing nucleic acid and detection method of nucleic acid
A nucleic acid and nucleic acid molecule technology, applied in the field of gene sequencing, can solve problems such as large fluctuations and instability, and achieve the effects of stable recovery, lower detection costs, and fewer purification steps.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0092] Example 1: Same-pipe reduction
[0093] In this embodiment, unless otherwise specified, the magnetic beads used are magnetic beads purchased from Axygen Corporation of the United States, also known as Axygen magnetic beads, and the product name is AxyPrep Magnetic Bead Purification Kits, Cat. No. MAG-PCR-CL-250. The beads are in suspension.
[0094] In this embodiment, unless otherwise specified, "room temperature" refers to 23°C±2°C.
[0095] In this embodiment, unless otherwise specified, "80% ethanol" refers to ethanol with a concentration of 80% by volume, also known as "80% V / V ethanol", which is composed of ethanol and NF-H 2 O (nuclease-free water, also known as Nuclease-Free Water) is prepared according to the volume ratio of 80:20.
[0096] In this example, TE buffer (TE Buffer) was purchased from Invitrogen, Cat. No. 12090015. The composition is as follows: 10 mM Tris-HCl (pH 8.0), 0.1 mM EDTA.
[0097] In this example, nucleic acid is oxidized and then ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com