Method for screening important characteristic genes related to drug resistance phenotype of bacteria based on machine learning
A feature gene, machine learning technology, applied in the field of gene sequencing
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0085] Embodiment 1 The design optimization of this application
[0086] As mentioned in the background technology section of this application, most of the existing research on bacterial drug resistance screening focuses on the screening of single nucleotide polymorphisms (SNPs), insertions / deletions (Indels) or k-mer features at the core genome level, However, in addition to these characteristics, it is also very important to screen for important non-core drug resistance genes associated with drug resistance phenotypes.
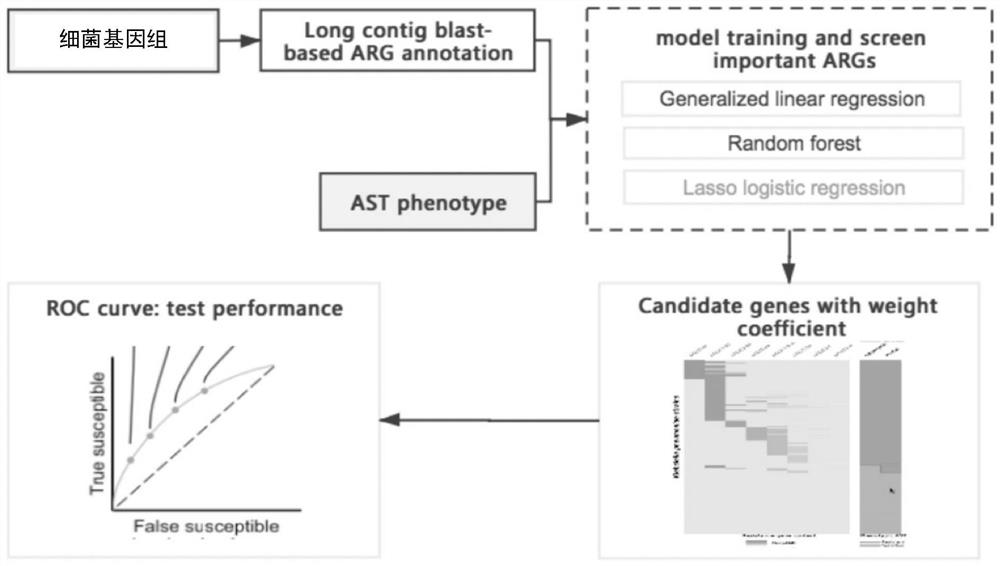
[0087] figure 1 This is the design idea of this application, based on this idea, this embodiment designs and optimizes specific methods for drug resistance screening. Take model selection as an example to show the establishment process of this application.
[0088] When performing the association analysis between genotype and drug sensitivity result data, this application compared and used GLM generalized linear model (R language glm(), stepAIC() command...
Embodiment 2
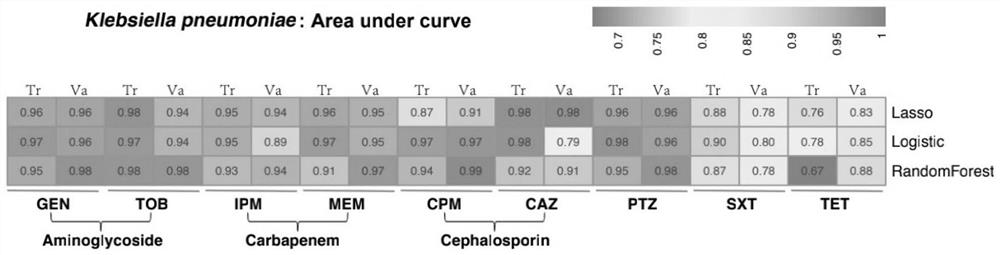
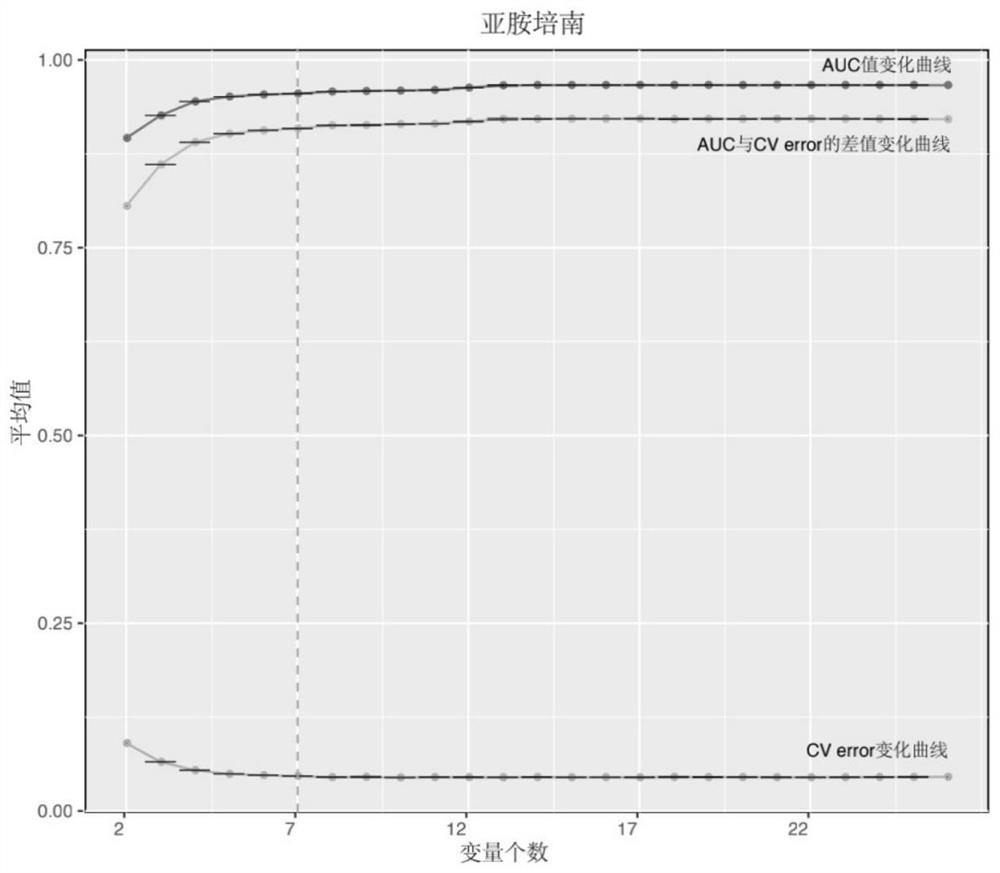
[0097] Example 2 Screening and verification of Klebsiella pneumoniae-resistant carbapenem and cephalosporin-related traditional Chinese medicine characteristic genes
[0098] Step 1. Search and download the Klebsiella pneumoniae strain genome and its corresponding antibiotic susceptibility test result data from the public database.
[0099] Download from the NCBI NDARO database: Open the website https: / / www.ncbi.nlm.nih.gov / pathogens / isolates, enter "Klebsiella pneumoniae" in the search bar to retrieve the information of Klebsiella pneumoniae, and then in the Matched Isolates sub-window, click "Choose columns" select "AST pheotypes" to display this column information, then download the tabular data of the entire window, sort out the lung strains with drug susceptibility test results data, according to the Assembly ID information, from the genome database of NCBI (ftp: / / ftp.ncbi.nlm.nih.gov / genomes) to download genome sequences in bulk.
[0100] Download from the PATRIC platf...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com