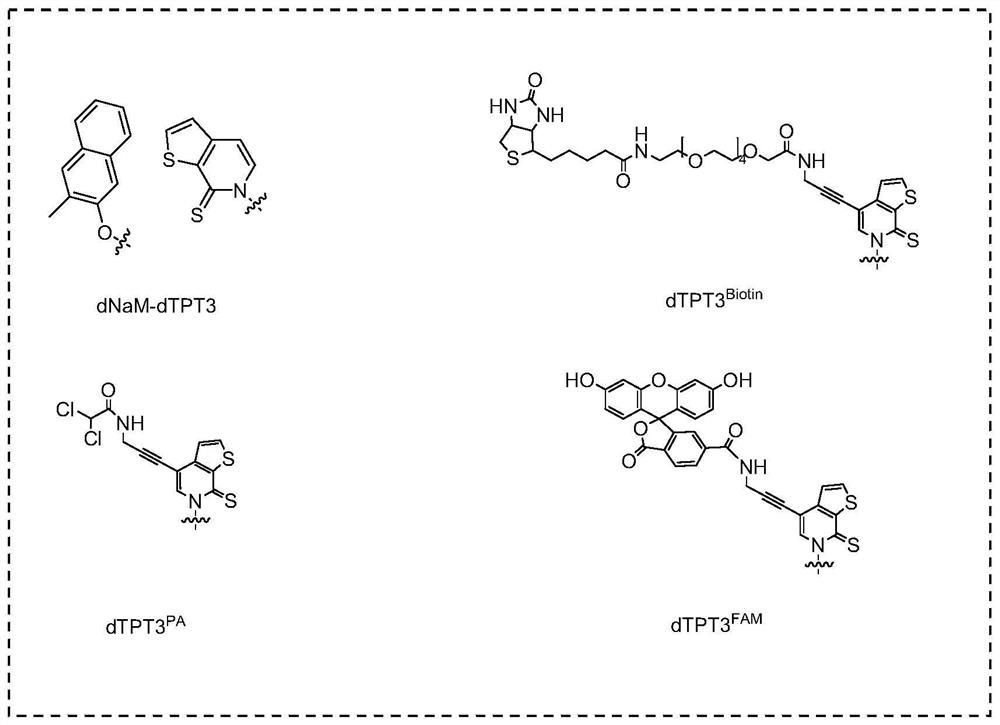
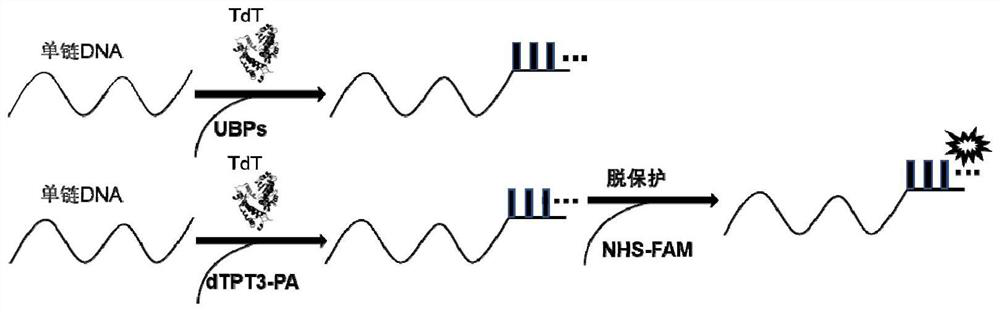
Method for template-free synthesis of non-natural base-containing oligonucleotide chain by using terminal deoxyribonucleotide transferase and application of non-natural base-containing oligonucleotide chain
A deoxyribonucleotide and base oligonucleotide technology, applied in biochemical equipment and methods, genetic engineering, DNA preparation, etc., can solve the problem of unresearched oligonucleotide chains containing non-natural bases, etc. problem, to achieve the effect of high coupling efficiency, low cost and convenient operation
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
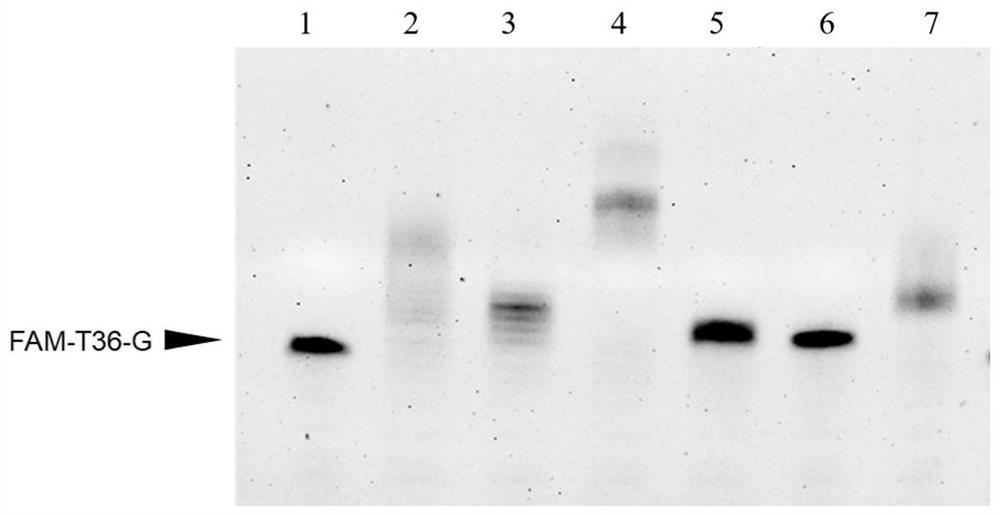
[0052] Preparation of oligonucleotide chains with unnatural base as dATP
[0053] (1) Mix 500nM priming chain single-chain FAM-T36-G, 1×TdT reaction buffer, the pH of TdT reaction buffer is 7.4, and 10mM magnesium acetate solution to obtain mixed solution 1;
[0054] (2) Add 50 μM dATP to the mixed solution 1 in step (1) to obtain the mixed solution 2;
[0055] (3) Add 1U / μL enzyme TdT to the mixed solution 2 described in step (2) to obtain the mixed solution 3;
[0056] (4) Incubate the mixed solution 3 in step (3) at 37° C. for 15 minutes, then heat-treat at 75° C. for 20 minutes to inactivate the enzyme TdT, and obtain the mixed solution 4;
[0057] (5) Add 2×TBE-Urea loading buffer to the mixed solution 4 in step (4), heat treatment at 95°C for 10 minutes to completely denature the oligonucleotide chain; analyze the enzyme TdT by denaturing gel The effect of adding an unnatural base to the 3' end of an oligonucleotide strand.
Embodiment 2
[0059] To prepare an oligonucleotide chain whose unnatural base is dTTP, the steps are the same as in Example 1, wherein dTTP is added in step (2), and the incubation time in step (3) is 30 minutes.
Embodiment 3
[0061] To prepare an oligonucleotide chain whose unnatural base is dGTP, the steps are the same as in Example 1, wherein dGTP is added in step (2).
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com