Nucleic acid metabolic enzyme activity detection method
A technology for nucleic acid metabolism and detection methods, applied in biochemical equipment and methods, microbial measurement/inspection, etc., can solve the problems of limiting the range of nuclease analysis, long experiment time, and small throughput, and achieve accurate activity detection results , High sensitivity, quick operation effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 2
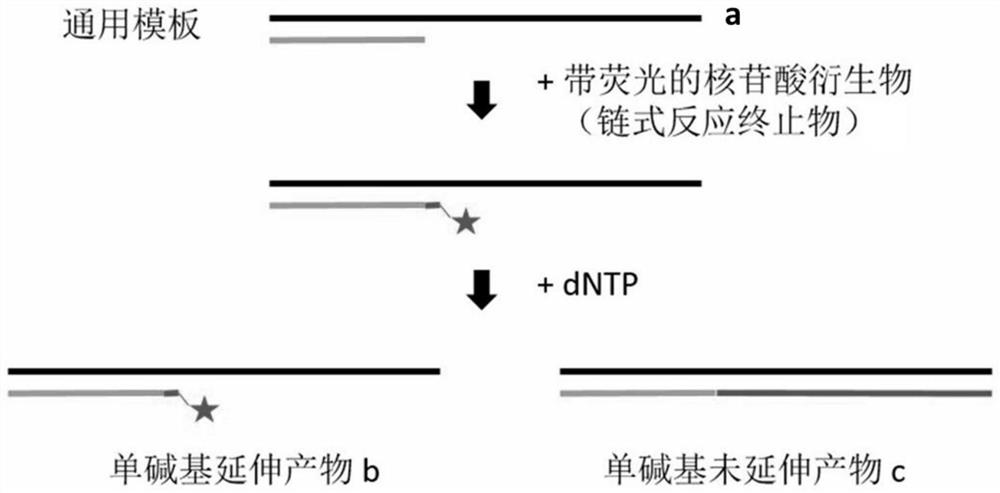
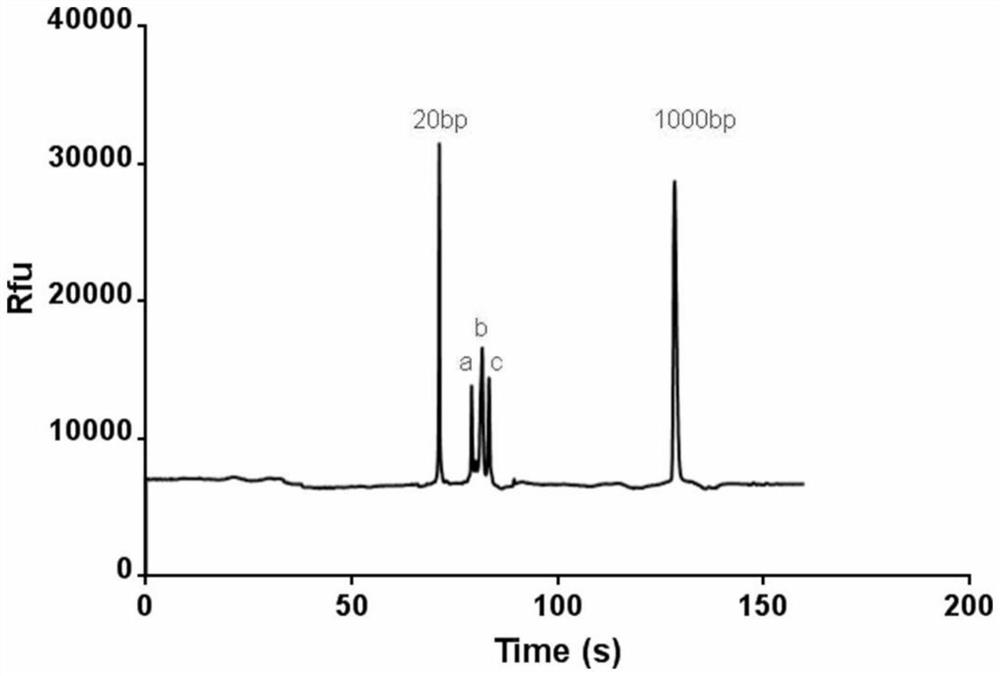
[0037] Single base extension error assay (capillary electrophoresis assay)
[0038] Design and synthesis of primers and templates:
[0039] Single-stranded nucleic acid sequence (5'-3'):
[0040] GACCGCGACTCCAGCCCTCTTACACCCAGTGGAGAAGCTCCCAACCAAGCTCTCTTGAGGATCTTGAAGGAAACTGAATTCAAAGTCGTCGCGGGATCA (SEQ ID NO. 1)
[0041] Primer sequence (5'-3'):
[0042] TGATCCCGCGACGACT (SEQ ID NO.2)
[0043] TGATCCCGCGACGACTTT (SEQ ID NO.3)
[0044] TGATCCCGCGACGACTTTG (SEQ ID NO.4)
[0045] TGATCCCGCGACGACTTTGAATT (SEQ ID NO. 5)
[0046]It is used to detect the reaction ability of different nucleic acid metabolizing enzymes and different fluorescently modified nucleotide derivatives, adding a general template and one of four different fluorescently labeled nucleotides (wrong nucleotides that do not match the template) into the reaction system Within 1 min, for the extension reaction, start the reaction by adding the nucleic acid metabolizing enzyme whose activity is to be detected to the...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com