Simple RNA library building method
A simple and reverse transcription technology, applied in the field of simple RNA library construction, can solve the problems of increasing the difficulty and time-consuming of RNA library construction, and achieve the effects of low cost, simple operation and small RNA loss.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
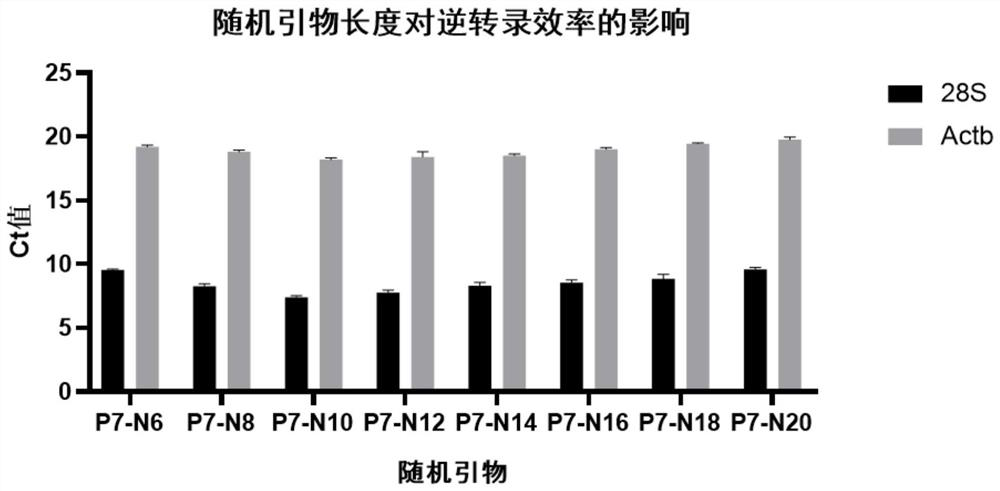
[0032] Example 1: Effect of Random Primer Length on Reverse Transcription Efficiency
specific Embodiment approach
[0033] In this example, we verified the effect of 6-20nt random primer length (No. 1-8 in Table 1) on reverse transcription efficiency. The specific implementation is as follows:
[0034] Table 2
[0035]
[0036] 94°C for 2min, 94°C-25°C for 0.1°C / s, and store at 4°C.
[0037] table 3
[0038] components Dosage The above reaction system 16μL 0.1M DTT 2μL SuperScript TM IIIRT (200U / μL, ThermoFisher)
1μL SUPERase·In TM RNase inhibitor (20U / μL)
1μL Total 20 μL
[0039] 10min at 25°C, 15min at 42°C, 5min at 85°C, and store at 4°C. Actin and 28S RNA were quantitatively analyzed by qPCR, and the quantitative primers are listed in Table 1.
[0040] The result is as figure 1 As shown, random primers with 8-14 bases have better reverse transcription efficiency.
Embodiment 2
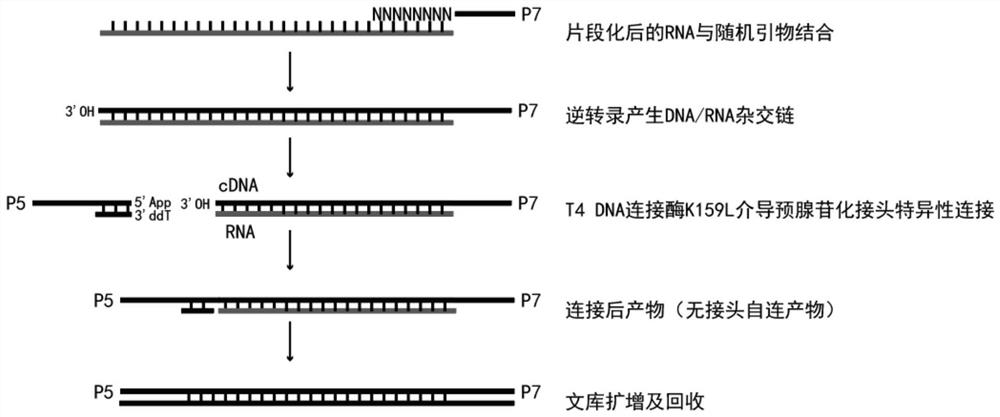
[0041] Example 2: Establishment of a simple RNA library construction process.
[0042] In this example, we established a simple RNA library construction process. The specific implementation is as follows:
[0043] P5 joint annealing: Dissolve 100μM P5-1 and 100μM P5-2 in 1×Annealing buffer (10mM Tris-HCl, 50mM NaCl, 1mM EDTA, pH 7.9), pipette equal volume and mix, 94℃ for 5min, 94-15℃ 0.1°C / min, store at 15°C. After annealing, the linker was diluted to a final concentration of 10 μM.
[0044] 1) RNA fragmentation and ribosomal RNA removal:
[0045] Table 4:
[0046] components Dosage RNA 10pg-1μg 0.3mM P7-N10 1μL rRNA Reverse Transcription Blocking Probe (202110257924.X) 1μL 5×First-Strand Buffer 2μL 10mM dNTPs 1μL Add DEPC water to 7μL
[0047] 5×First-Strand Buffer: 250mM Tris, 375mM KCl, 15mM MgCl 2 , pH 8.3.
[0048] 94°C for 10min, 75°C for 1min, 55°C for 1min, and store at 4°C.
[0049] 2) RNA reverse tran...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com