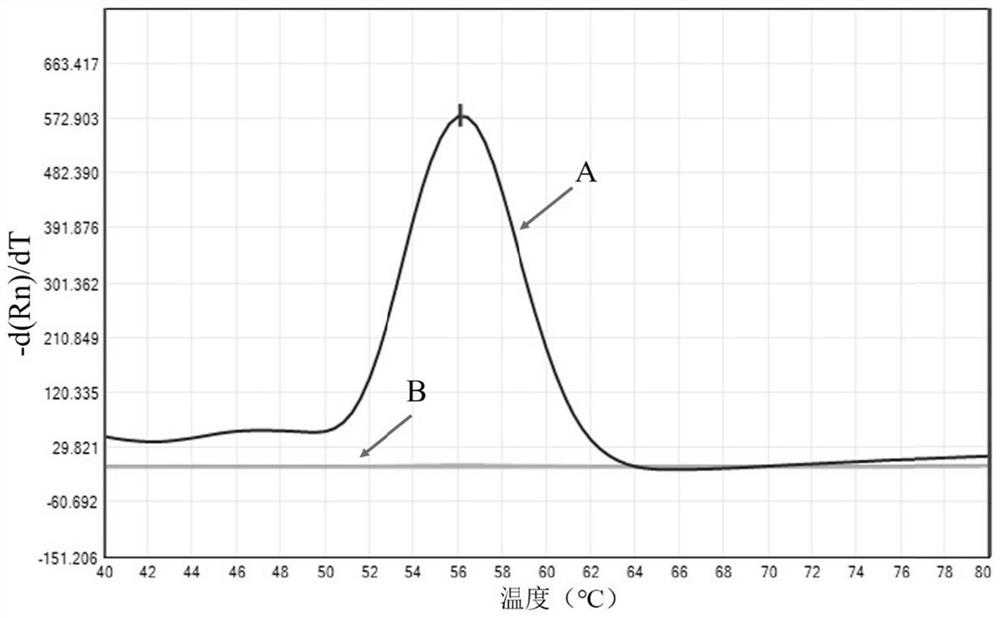
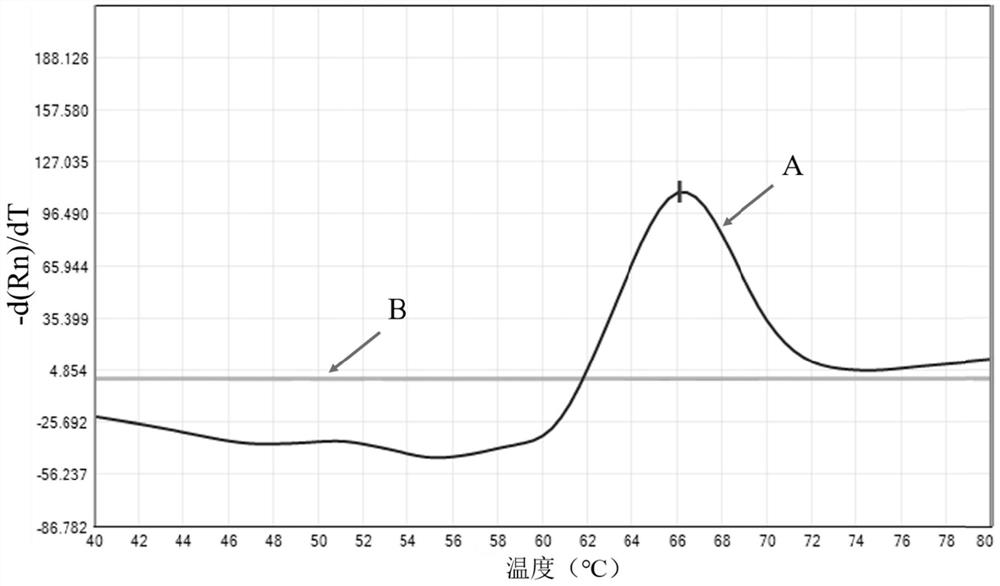
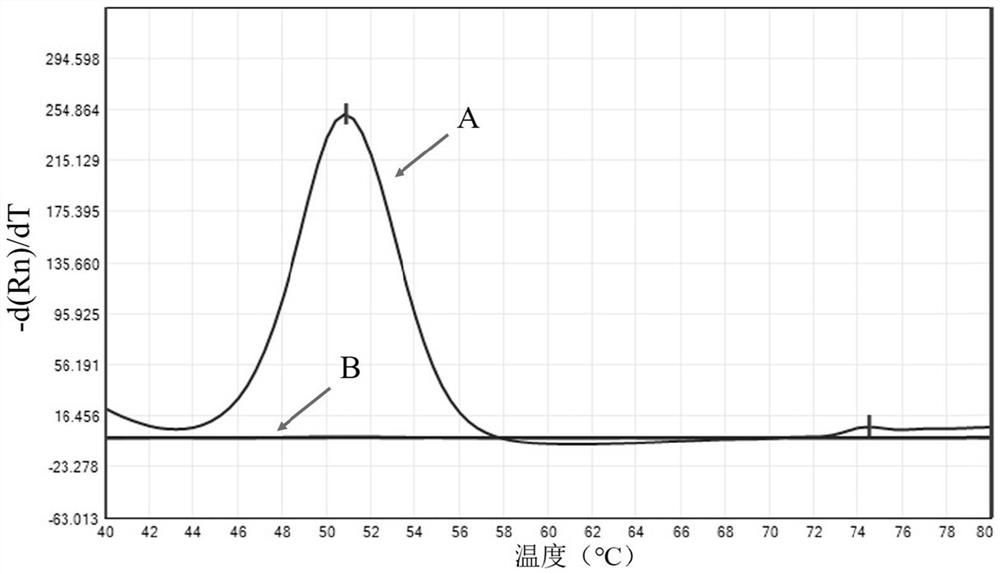
Kit for detecting nontuberculous mycobacteria, detection method and application thereof
A detection kit and technology of mycobacteria, applied in the field of molecular biology, can solve the problems of different clinical manifestations, difficulties in identification and differentiation, etc., and achieve the goals of shortening detection time, predicting disease risk and health management, and improving detection throughput Effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0098] Example 1 Design and synthesis of primers and probes
[0099] According to the Mycobacterium avium complex, Mycobacterium abscessus and Mycobacterium fortuitum among the non-tuberculous mycobacteria published by Genebank, the minor species include Mycobacterium kansasii, Mycobacterium chelonis, Mycobacterium goldenscens, Mycobacterium toadiensis, Mycobacterium simianum, Mycobacterium sulja, Mycobacterium marmoe, Mycobacterium haemophilus, Mycobacterium intracellulare, Mycobacterium scrofula, Mycobacterium asiatica and human internal standard The genome sequence of the gene, compare and analyze the differences of each genome sequence, and design probes and primers.
[0100] The final designed primer pairs and corresponding probe sequences are as follows:
[0101] Primer pairs for NMT:
[0102] Upstream primer NMT-F: 5'-AGAAGACCGACGACGTBGC-3' (SEQ ID NO: 1);
[0103] Downstream primer NMT-R: 5'-RTCTSGGTGAYCTTYTCSAC-3' (SEQ ID NO: 2);
[0104] Probe sets for NMT:
[0...
Embodiment 2
[0121] Example 2 A fluorescent quantitative PCR-HRM detection kit for non-tuberculous mycobacteria
[0122] The components of the non-tuberculous mycobacteria fluorescent quantitative PCR-HRM detection kit in this embodiment are as shown in Table 3 and Table 4:
[0123] Table 3 Components of Kit 1
[0124]
[0125] Table 4 Components of Kit 2
[0126]
[0127] Note: After the plasmid was dissolved with 1×TE buffer, the concentration was determined by UV spectrophotometry, and then diluted to 1×10 with 1×TE buffer. 5 copies / μL.
[0128] Wherein, PCR reaction systems 1 and 2 include the NMT primer pair and NMT probe set in Example 1, as well as human internal standard gene primers and probes.
[0129] Wherein, the nucleotide sequence of the human internal standard gene primer is:
[0130] Upstream primer: human internal standard gene F: 5'-TAGGGAGTATATAGGTTGGGGAAGTT-3' (SEQ ID NO: 10);
[0131] Downstream primer: human internal standard gene R: 5'-AACACACAATAACAAACACA...
Embodiment 3
[0148] Example 3 Preparation of standard positive samples and PCR-HRM detection
[0149] (1) Extraction of DNA in standard positive sample nucleic acid:
[0150] The samples to be tested were Mycobacterium avium, Mycobacterium abscessus and Mycobacterium fortuitum, and the secondary species were Mycobacterium kansasii, turtle clam Mycobacterium chelonae, Mycobacterium gordonae, Mycobacterium xenopi, Mycobacterium simiae, Mycobacterium szulgai, Malmo Mycobacterium malmoense, Mycobacterium haemophilum, Mycobacterium intercelleulare, Mycobacterium scrofulaceum, and Mycobacterium arisiense (approximately 2.0 ×10 5 copies / mL) and human internal standard gene.
[0151] Reagents and instruments for DNA extraction of samples to be tested: Nucleic acid extraction was performed using the magnetic bead method nucleic acid extraction or purification kit of Guangzhou Weijie Biomedical Technology Co., Ltd. The nucleic acid extraction instrument is Weijie Auto-Extractor32 nucleic acid ex...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com