Library construction method based on promoter and library thereof
A promoter and library technology, applied in the field of gene sequencing, can solve the problems of inapplicability and low coverage of single-cell methylation sequencing, and achieve the effect of improving the coverage.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
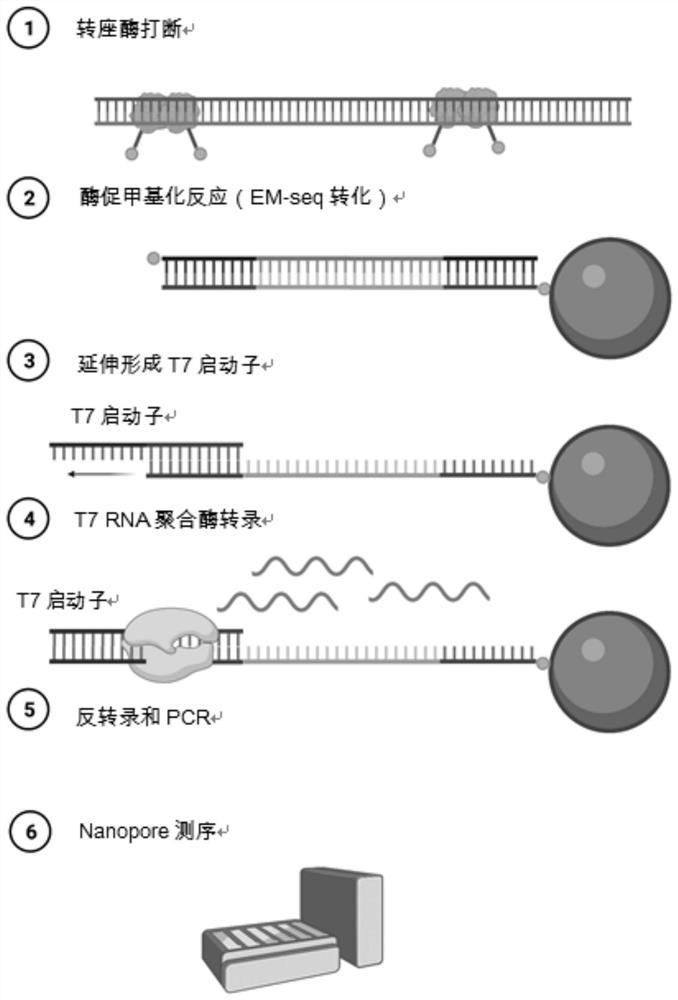
[0090] figure 1 It is a schematic diagram of the database construction process in this embodiment.
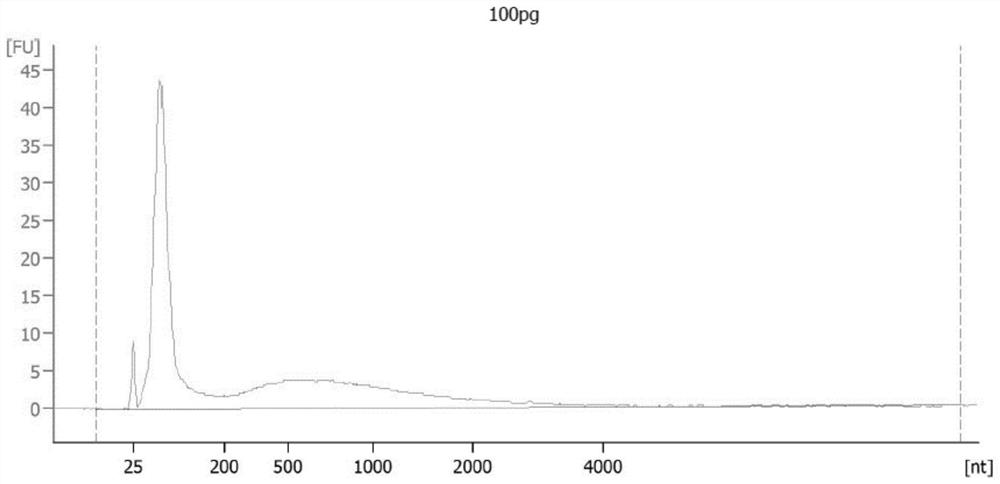
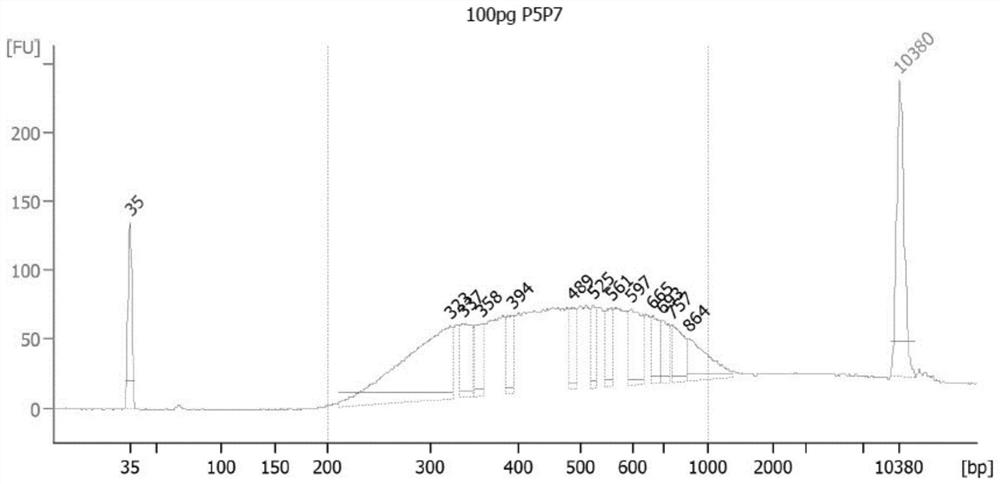
[0091] In this example, a 100 pg DNA methylation library was constructed.
[0092] Specific steps are as follows:
[0093] 1. Annealing of oligo (New Tn5-pho) and oligo (New bio-bottom) to form a double-stranded linker, and then embedding with transposase Tn5 to form a complex. "oligo" refers to an oligonucleotide, "pho" refers to a phosphate group, and "bio-bottom" refers to an oligonucleotide modified with biotin.
[0094] The nucleotides used in this example are shown in Table 2.
[0095] Table 2
[0096]
[0097] "N" represents a random base, which is any one of A, T, C, and G.
[0098] 2. Tn5 interrupts genomic DNA
[0099] Take 100pg of DNA and interrupt it with the Tn5 enzyme embedded in the linker. The interrupted reaction system is 10 μL, in which the amount of transposase used is 0.5 μL, and the amount of 5XtagH buffer used is 2 μL. React on a PCR instrument at...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com