Construction body of nano antibody S43 and application thereof
A nanobody and pentamer technology, applied in the direction of antibodies, applications, antiviral agents, etc., can solve the problems of ineffective reduction of lung viral load, low drug concentration, and reduced antiviral effect of neutralizing antibodies
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
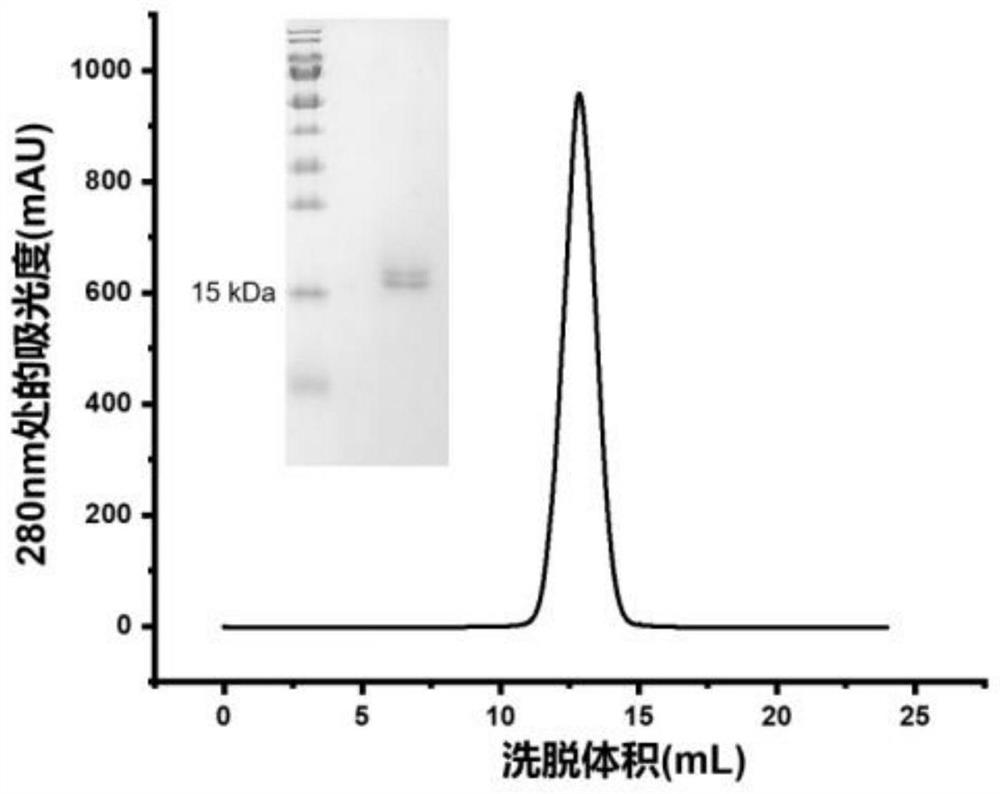
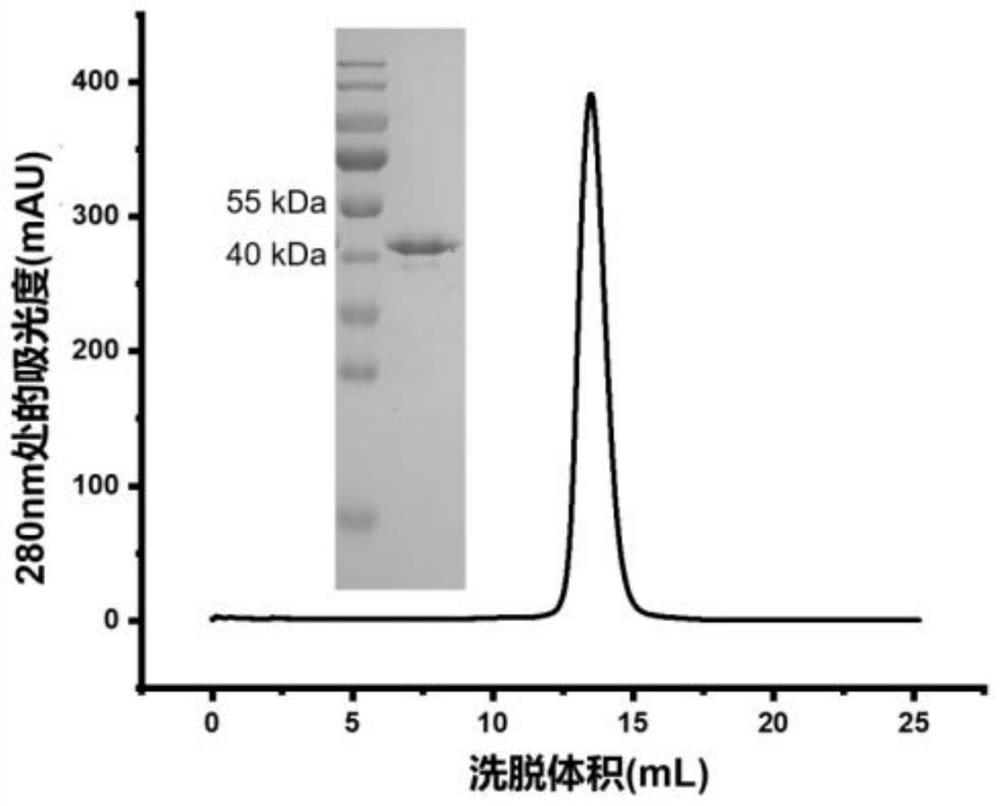
[0085] Example 1: Construction, expression and purification of antibodies based on trivalent form (TS43) and IgM pentamer form (MS43) of Nanobody S43
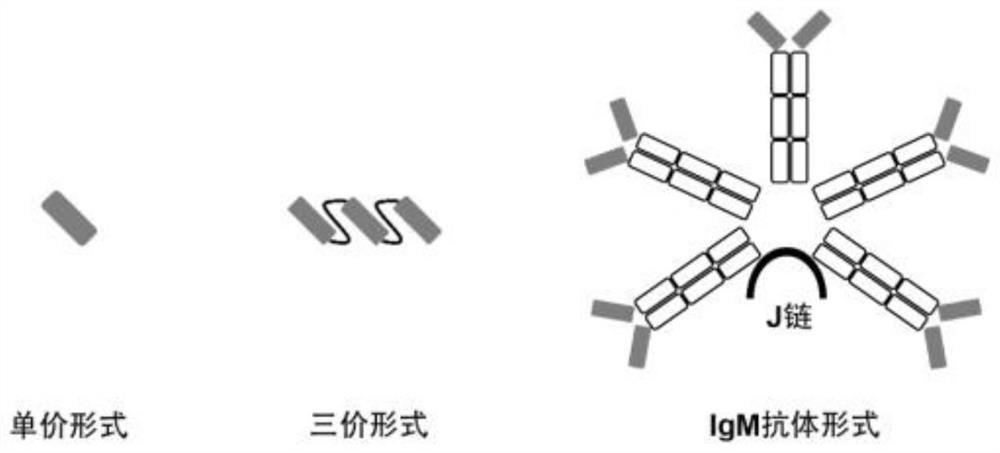
[0086] The structural schematic diagrams of the monovalent Nanobody and its trivalent form and IgM pentamer form in this example are as follows: figure 1 shown.
[0087] The basic nanobody S43 used is obtained by the laboratory by immunizing alpacas with SARS-CoV-2 S protein, constructing an antibody library, and screening by phage display technology; the amino acid sequence of monovalent nanobody S43 is shown in SEQ ID NO: 8. showed that it can bind SARS-CoV-2 RBD with high affinity and specificity (binding constant is 1.2E-10 ± 1.4E-11M), and in pseudovirus neutralization experiments, it can neutralize SARS-CoV-2 with high neutralizing activity. CoV-2 pseudovirus, all of which show that the S43 nanobody is a novel coronavirus (SARS-CoV-2) alpaca-derived nanobody that can bind to SARS-CoV-2 RBD with high affinity and has hig...
Embodiment 2
[0091] Example 2: Expression and purification of SARS-CoV-2 and its variant strain RBD
[0092] The coding sequence of 6 histidine tags (hexa-His-tag) is connected to the 3' end of the coding sequence of the RBD protein of the original strain of SARS-CoV-2 (its amino acid sequence is shown in SEQ ID NO: 18) and the translation stop codon TGA, through the restriction endonuclease sites EcoRI and XhoI, it was constructed into the pCAGGS vector (purchased from Invitrogen), and then the resulting recombinant vector was transfected into 293F cells (purchased from Invitrogen) for Expression of SARS-CoV-2 RBD-his protein.
[0093] The coding sequence of the RBD protein (its amino acid sequence is shown in SEQ ID NO: 19) of the SARS-CoV-2 variant strain Omicron (B.1.1.529) subtype BA.1 and Omicron (B.1.1.529) ) The coding sequence of the RBD protein of subtype BA.2 (its amino acid sequence is shown in SEQ ID NO: 20) is connected to the coding sequence and translation of 6 histidine t...
Embodiment 3
[0095] Example 3: Surface plasmon resonance technology detects the binding ability of each antibody to the RBD protein of the original SARS-CoV-2 strain and its variants
[0096] Surface plasmon resonance analysis was performed using a Biacore 8K (Biacore Inc.). Specific steps are as follows:
[0097] The monovalent Nanobody S43 and its constructs TS43 and MS43 prepared in the above examples were biotinylated, and then immobilized on the Series Sensor Chip SA chip (Cytiva Life Sciences); with PBST buffer (2.7 mM KCl, 137 mM NaCl) , 4.3mM Na 2 HPO 4 , 1.4mM KH 2 PO 4 , 0.05% Tween) The RBD protein of the original SARS-CoV-2 strain and its variant strains prepared in the above-mentioned examples were diluted by multiple ratios, and the samples were loaded to the chip one by one from low concentration to high concentration. Calculation of binding kinetic constants was performed using BIAevaluation software 8K (Biacore, Inc.). Equilibrium dissociation constant (K) between ea...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com