Method for improving linear amplification detection accuracy through multiple detection, kit and application
A linear amplification and multiple detection technology, applied in biochemical equipment and methods, microbial measurement/inspection, etc., can solve the problems of undetectable, limited error, and difficult detection of samples with severe cell heterogeneity, and achieve The effect of reducing missed detection of target DNA molecules, reducing copy number detection bias, and avoiding primer bias errors
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
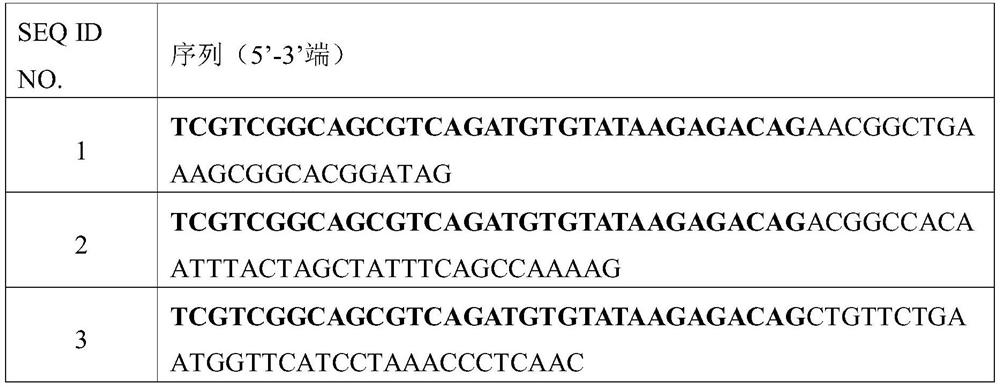
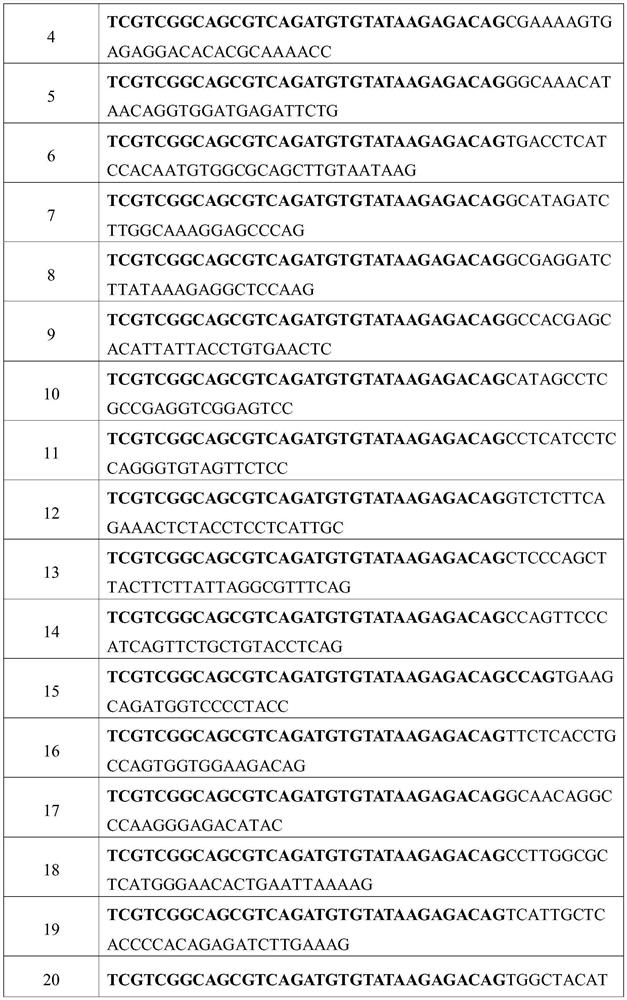
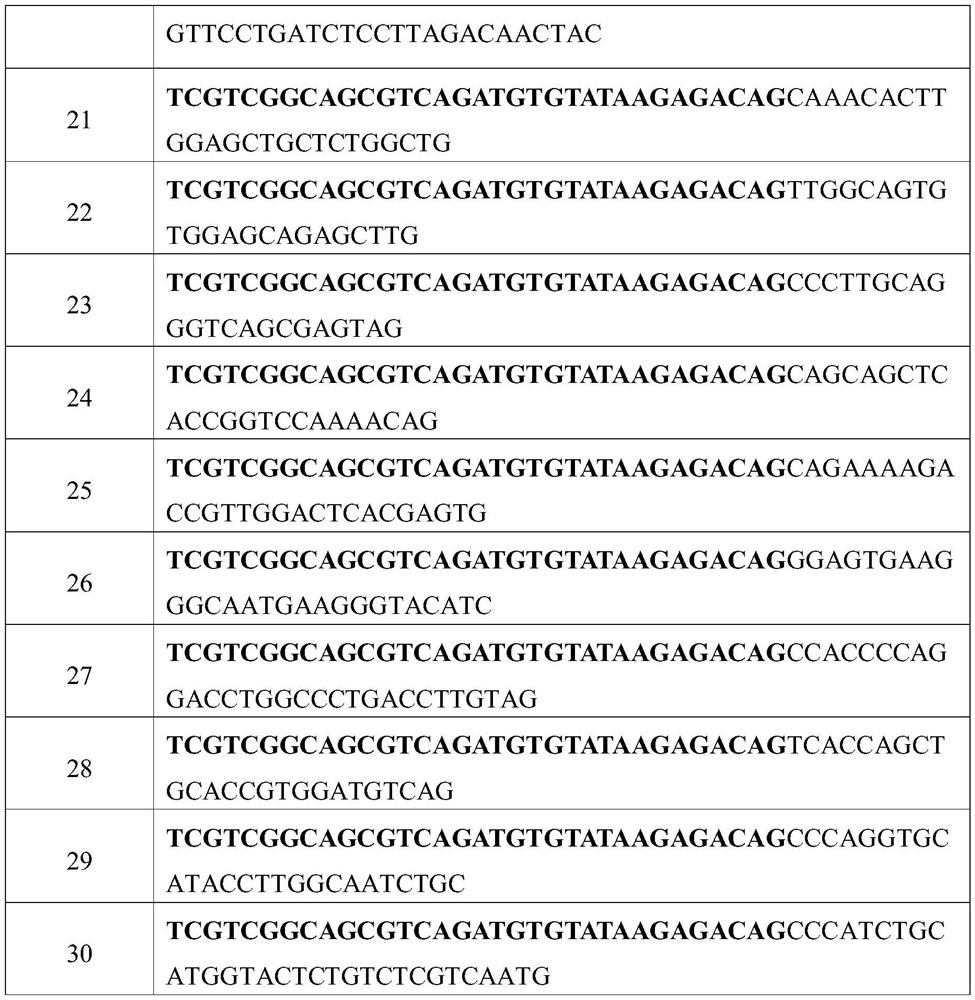
[0098] Example 1: Effects of Different Primer Quantities and Different Normalization Algorithms on the Accuracy of Linear Amplification Detection of CNV 1. Linear Amplification
[0099] 1.1 Linear amplification system preparation, as shown in Table 6:
[0100] Table 6. Linear Amplification System
[0101]
[0102]
[0103] 1) According to the above table 6, firstly prepare 5×Apo Buffer, APO biotin-dNTP mix 1 and Apo-Enchanted DNA polymerase I into a Mix solution;
[0104] 2) Subsequent addition of primer combinations and detection samples / control samples, adding water to make the reaction volume of each sample 20 μL.
[0105] 3) Start the linear amplification program.
[0106] 4) The linear amplification product was purified by magnetic bead method.
[0107] 1.2 Linear amplification condition settings, as shown in Table 7:
[0108] Table 7. Linear Amplification Conditions
[0109]
[0110] 2. Linker Ligation Reaction
[0111] 2.1 The joint connection system prep...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com