Method for producing peptides containing non-natural amino acids
A technology of unnatural amino acids and amino acids, applied in the field of producing peptides containing unnatural amino acids, can solve problems such as reducing fidelity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0292] Example 1. Construction of E. coli strains
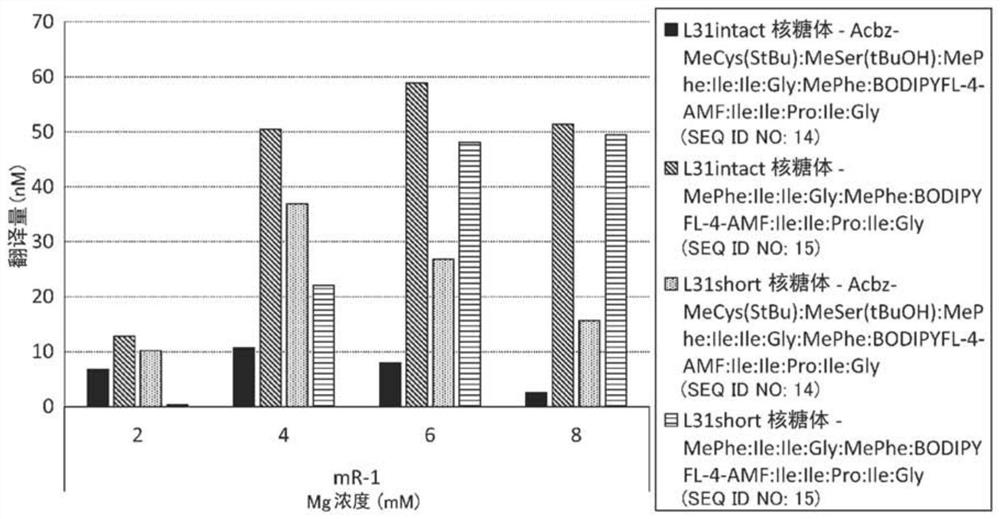
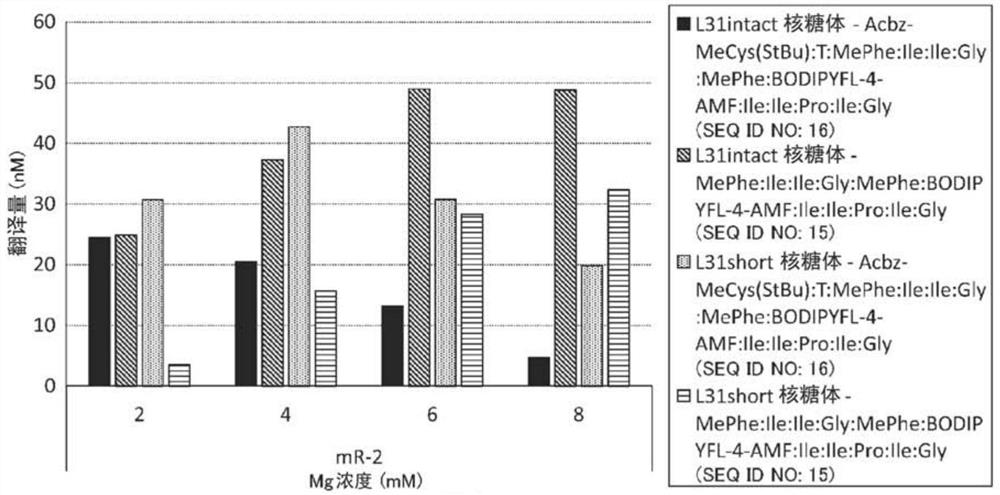
[0293] A strain lacking ompT (Protease 7) (defined as an L31intact strain) and an L31short strain expressing the 1st to 62nd amino acids of the N-terminal of the L31 protein were constructed. During ribosome purification, protease 7 is known to degrade between amino acids 62 and 63 of the L31 protein.
[0294] E. coli strains were constructed according to the procedure of Quick and Easy Conditional Knockout Kit (loxP / Cre) (Gene Bridges).
[0295] Preparation of functional cassettes with added homology arms
[0296] PCR reactions were performed using PrimeSTAR HS DNA polymerase (Takara Bio Inc., R010A) using the cassette loxP-PGK-gb2-neo-loxP (Gene Bridges, A003) as template. The obtained PCR product was purified using the QIAquick PCRPurification Kit (QIAGEN, 28104). The concentration of the functional cassette is approximately 200 ng / μL. PCR was performed using Oligol (SEQ ID NO: 3) and Oligo2 (SEQ ID NO: 4) as primers...
Embodiment 2
[0299] Example 2. Method for preparing ribosomes
[0300] Culture of Escherichia coli
[0301] The W3110 strain (WT) was grown.
[0302] The bacterial cells of the W3110 strain were inoculated into the pre-medium (glycerol 5g / L, yeast extract 6g / L, KH 2 PO 4 4g / L, K 2 HPO 4 9.3g / L) and pre-incubated. The precultured bacterial cells were added to 30L main medium (glycerol 10g / L, yeast extract 10g / L, polypeptone N15g / L, KH 2 PO 4 4g / L, MgSO 4 ·7H 2 O 2.4g / L, FeSO 4 ·7H 2 O 0.04g / L, CaCl 2 ·2H 2 O 0.04g / L, decyl alcohol LG-1090.24g / L), make OD 600 is 0.1. Cultures were performed in 50-L culture vessels. Cells were incubated at 37°C for 5.4 hours, when the OD 600 Recycled when reaching 33.5. The culture solution was aliquoted into 500 mL and left at room temperature for 1 hour and at 4°C for another hour. Thereafter, the mixture was centrifuged at 6000×g for 10 minutes, the pellet was suspended in D-PBS(-) (Takara Bio Inc., T9181), and then centrifuged aga...
Embodiment 3
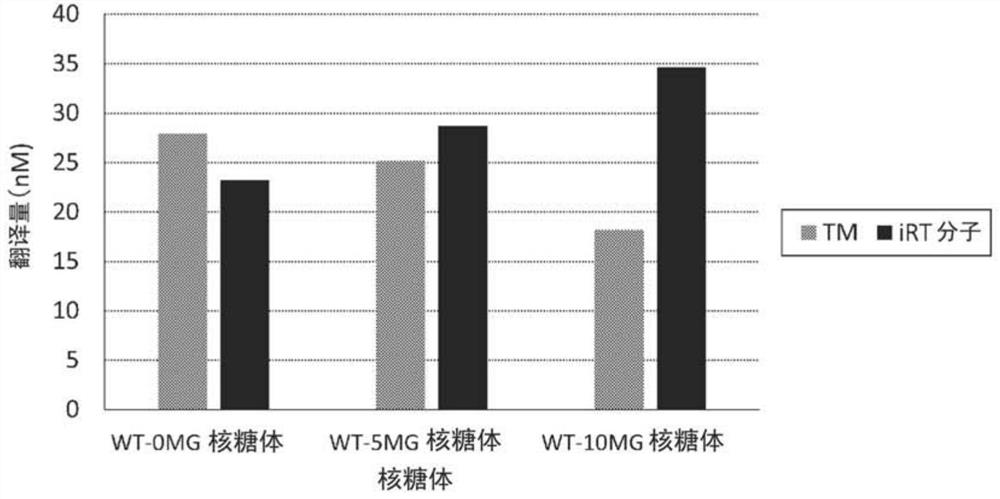
[0325] Example 3. Analysis of percent degradation of L31
[0326] The percent degradation of L31 in WT-10MG ribosomes, WT-5MG ribosomes, and WT-0MG ribosomes was analyzed.
[0327] Sample preparation method
[0328] Ribosomes (40 pmol) were diluted with 38 μL of water. 1% trifluoroacetic acid was added to precipitate ribosomal RNA. Centrifugation was performed and the supernatant was mixed 1:1 with matrix (50% acetonitrile, 5 mg / mL sinapic acid) and crystallized for MALDI / MS by spotting 1 [mu]L on the plate.
[0329] Analysis by MALDI / MS
[0330] Measurements were performed on a mass spectrometer (ABS CIEX·TOF / TOF 5800) using linear positive mode. Calibration was performed using ribosomal proteins as internal standards. Use properly calibrated measurements. The percentage of intact L31 was calculated by dividing the MS intensity of the intact L31 by the sum of the MS intensity of the intact L31 and the MS intensity of the short L31 (short L31). Measurements were mad...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com