(R)-2-octanol dehydrogenase, process for producing enzyme, DNA encoding enzyme and process for producing alcohol by using same
A dehydrogenase, octanol technology, applied in biochemical equipment and methods, botanical equipment and methods, applications, etc., can solve problems such as unfavorable industrial applications, chemically unstable NADPH, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
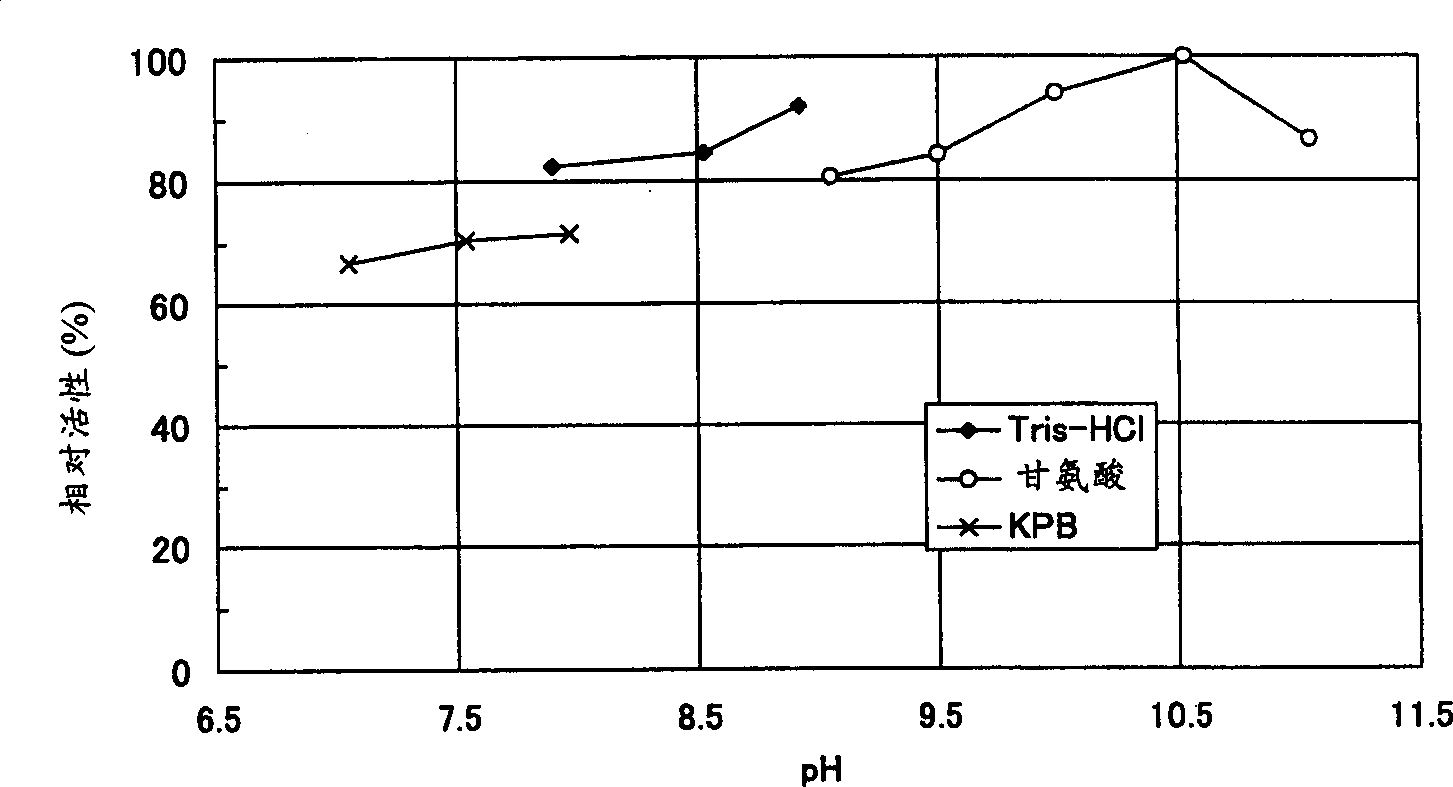
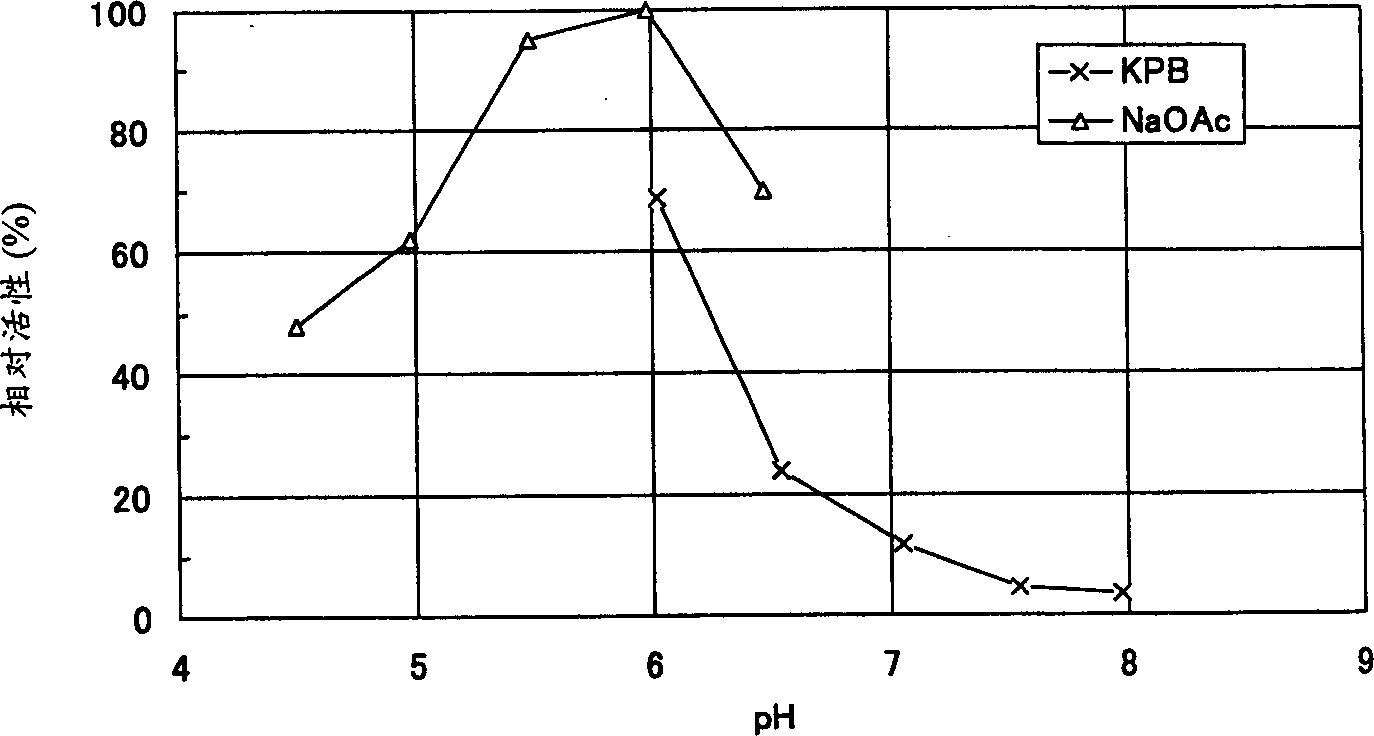
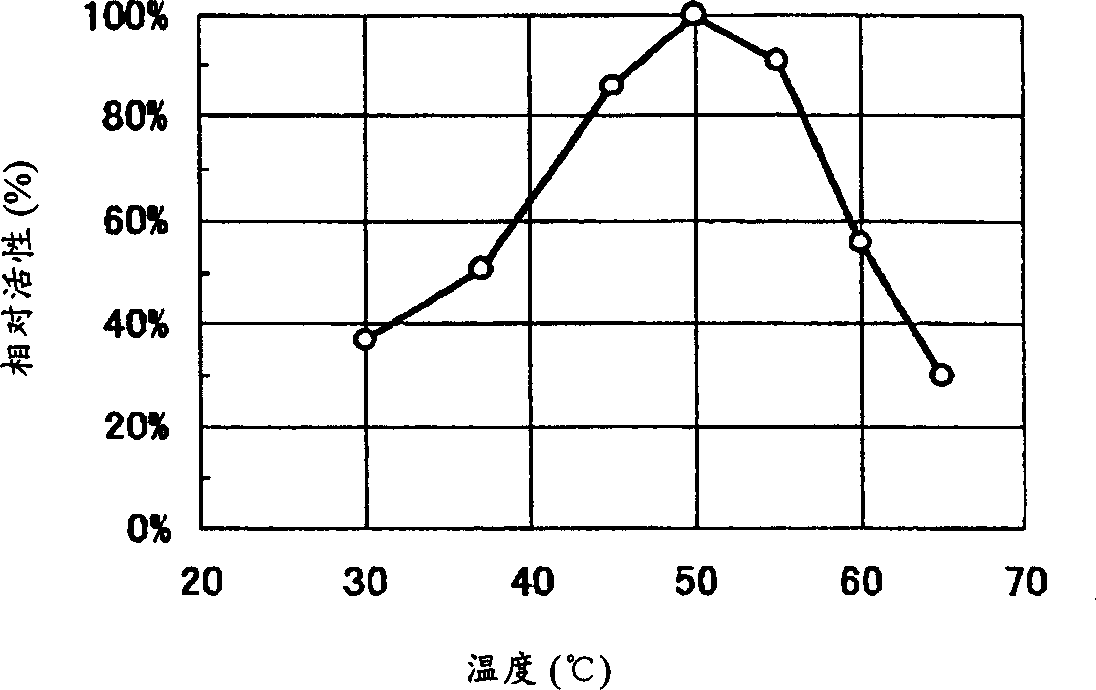
Image
Examples
Embodiment 1
[0194] The present invention is illustrated in detail with reference to the following examples, but is not limited thereto. Example 1: Screening of (R)-2-octanol dehydrogenase
[0195] Yeast (R)-2-octanol dehydrogenase is screened by activity staining method after electrophoresis. The substrate is (R)- or (S)-2-octanol substrate. The reaction mixture includes NAD+, phenazine sulfate Dimethyl ester (PMS), nitro blue tetrazolium (NBT). The composition of the reaction mixture is as follows:
[0196] 5mM (R)- or (S)-2-octanol
[0197] 1.3mM NAD+
[0198] 0.13mM PMS
[0199] 0.48m MNBT
[0200] As a result, enzymes that were not stained when the substrate was (S)-2-octanol but preferentially stained when the substrate was (R)-2-octanol were found in the following yeast strains. These enzymes with (R)-2-octanol dehydrogenase activity also have the activity of producing ethyl (S)-4-chloro-3-hydroxybutyrate by reduction of ethyl 4-chloroacetoacetate.
[0201] ●Pichia finlandica...
Embodiment 2
[0204] Ogataea wickerhamii: IFO 1706 Example 2: Culture conditions for production of (R)-2-octanol dehydrogenase
[0205] The strain producing (R)-2-octanol dehydrogenase in Example 1 was cultured in a carbon source where 2% glucose was changed to 2% glycerol or 1% methanol. It turned out that Pichia finlandica could induce this enzyme in all cultures. The enzyme was strongly induced by Candida utilis when methanol was used as the carbon source. Example 3: Purification of (R)-2-octanol dehydrogenase from Pichia finlandica
Embodiment 3
[0206] Pichia finlandica DSM 70280 strain was cultured in 4.8L medium A, and microbial cells were prepared by centrifugation. The obtained microbial cells were suspended in 100 mM Tris-HCl buffer (pH 8.0), 10% glycerol, 1 mM ethylenediaminetetraacetic acid 2Na (EDTA Na), 0.02% 2-mercaptoethanol, and 2 mM benzenemethanesulfonyl fluoride ( PMSF), homogenized with a glass bead stirrer (Biospec). Then, the microbial cell debris is removed by centrifugation to obtain a cell-free extract. Protamine sulfate was added, and the nucleic acid was removed by centrifugation to obtain a supernatant. Ammonium sulfate was added to the supernatant to give a precipitated fraction at 40-75% saturation. A standard buffer solution containing 30% saturated ammonium sulfate was added to the precipitated fraction to obtain an enzyme solution containing 40% (final concentration) saturated ammonium sulfate. The enzyme solution was centrifuged to obtain a supernatant. The supernatant was applied to ...
PUM
| Property | Measurement | Unit |
|---|---|---|
| optical purity | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com