Identification of DNA variant associated with adult type hypolactasia
An intestinal lactase, adult-based technology for the identification of DNA variants associated with adult-type intestinal lactase deficiency at low cost
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
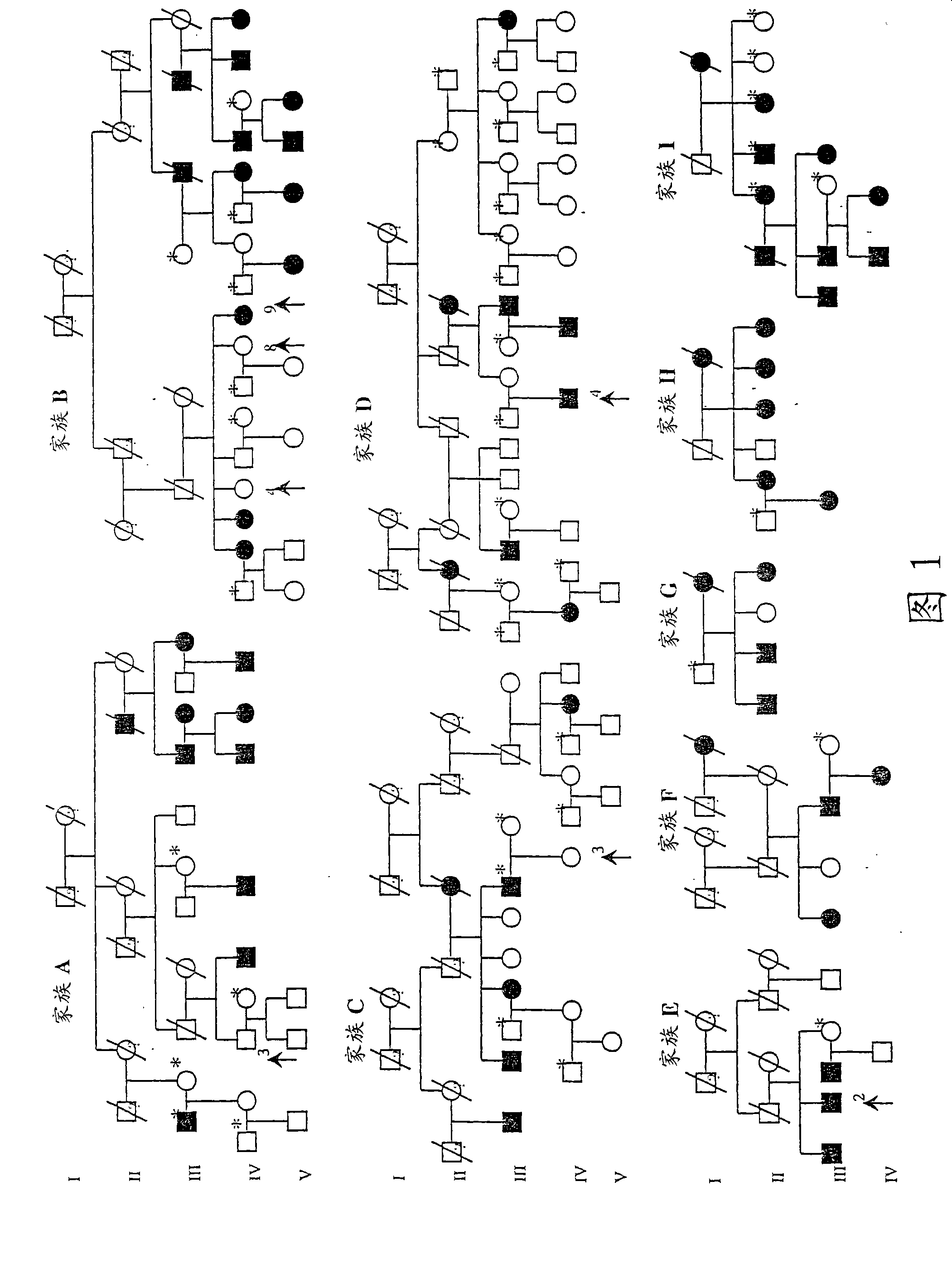
[0102] Example 1: Linkage and linkage disequilibrium analysis
[0103] Analysis of seven polymorphic microsatellite markers located between D2S114 and D2S2385 flanking the LPH gene on 2q21 in nine extended Finnish intestinal lactase deficiency families ( figure 1 ). Significant evidence of linkage to markers D2S314, D2S442, D2S2196 and D2S1334 was found, with the maximum log odds score value obtained for the D2S2196 marker being 7.67 at θ=0 (Table 1). Obligate recombination events are detected by the marker D2S114 (family B, IV3), thereby determining the centromeric boundary of the lactase persistence / non-persistence site; obligate recombination events are detected by the marker D2S2385 (family B, IV17) ( figure 1 , Table 1), so as to determine the telomere boundary of this site. To pinpoint this critical region, nine additional polymorphic markers were analyzed (Table 1). Based on the detected linkage, the linkage disequilibrium (LD) of the region is monitored with allel...
Embodiment 2
[0105] Example 2: Extended Haplotype Analysis
[0106] In the first stage, ten highly polymorphic microsatellite markers flanking the LPH gene on 2q21 were analyzed as described elsewhere 40,55 . Briefly, using the following gene distances, the analysis from the Généthon ResourceCenter 55 Ten highly polymorphic microsatellite markers on 2q near the lactase gene: cen-D2S114-1cM-D2S1334-0cM-D2S2196-0cM-D2S442-2cM-D2S314-2cM-D2S2385-1cM-D2S2288-1cM-D2S397- 1cM-D2S150-1cM-D2S132. The sequence of most of the above markers is mainly obtained from chromosome 2 provided by the Généthon map (Chumakov et al., 1995 56 ) obtained from the physical YAC contigs. In a total volume of 15 μl containing 12ng template DNA, 5pmol primers, 0.2mM each nucleotide, 20mM TrisHCL (pH 8.8), 15mM (NH 4 ) 2 SO 4 , 1.5mM MgCl 2 , 0.1% Tween 20, 0.01% gelatin and 0.25 U Taq polymerase (Dynazyme, Finnzymes) for PCR. The 5' end of one of the primers consists of 32 P-γATP is radioactively labeled. T...
Embodiment 3
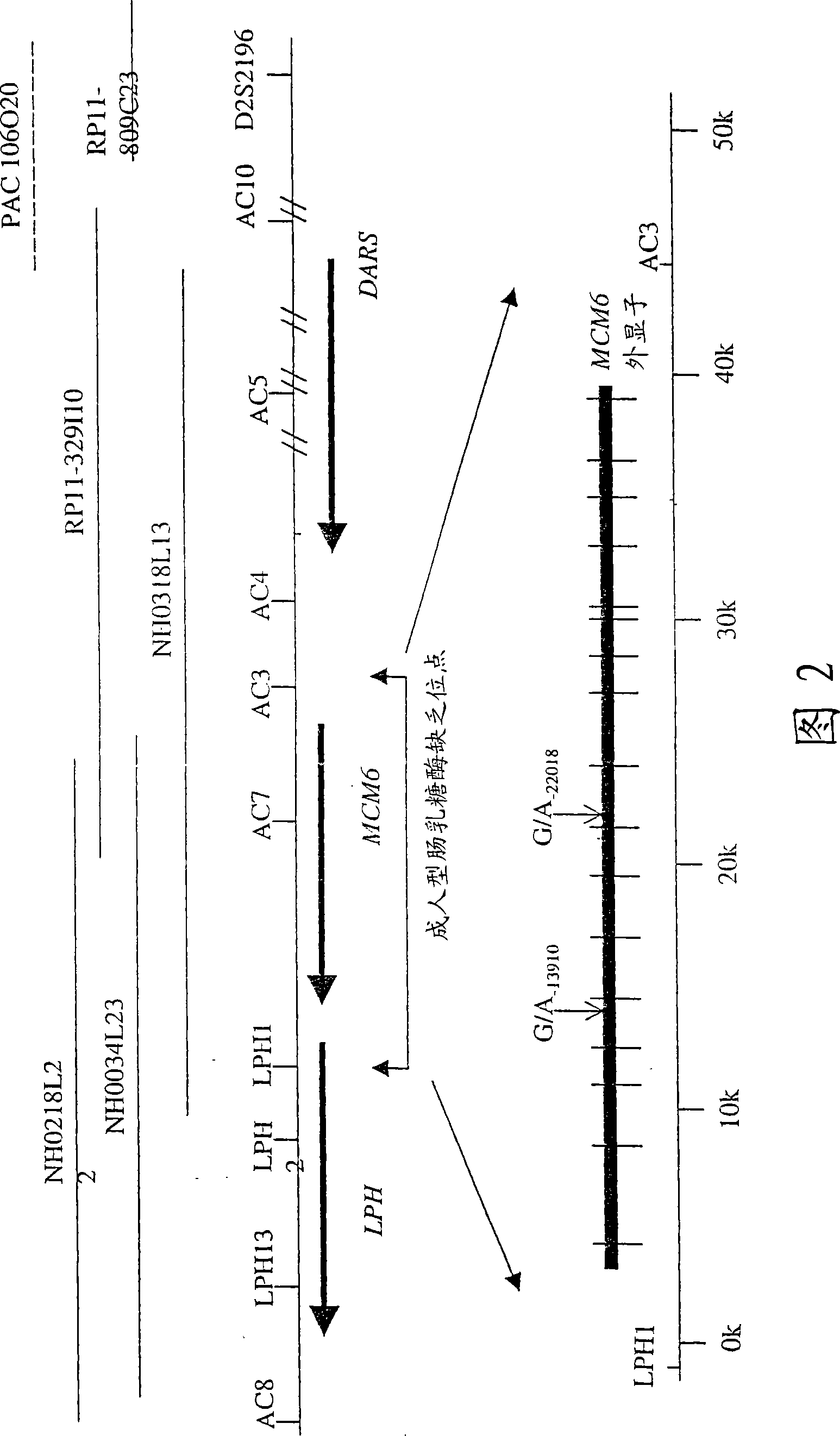
[0110] Example 3: Sequence analysis of adult-type intestinal lactase deficiency loci
[0111] A 47 kb region between markers LPH1 and AC3 was amplified and sequenced in overlapping PCR fragments of genomic DNA from some members of nine intestinal lactase-deficient families. This region includes the 36 kb minichromosome maintenance (MCM6) gene covering the critical 47 kb region 18 ( figure 2 ). Except for a total of 52 variants, no variations were detected in the coding region of the MCM6 gene; 43 SNPs and 9 deletion / insertion polymorphisms were identified in this critical 47 kb region (Table 2). Only two variants (C / T -13910 and G / A -22018 ) was associated with the lactase persistence / non-persistence trait in the Finnish family (Tables 2 and 3). First related variant C / T -13910 Within the intron 13 of the MCM6 gene, it is located at the -13910 site away from the first ATG codon of the LPH gene. The second related variant G / A -22018 Within the intron 9 of the MCM6 gen...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com