Signal detection method for gene chip of oligonucleotide with length more than 50 basic groups
A gene chip and oligonucleotide technology, applied in the biological field, can solve problems such as operator personal injury and environmental nuclear radiation pollution, and achieve the effects of high sensitivity, reliable results and high detection rate.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment
[0020] (1) Preparation of gene chips for oligonucleotides longer than fifty bases
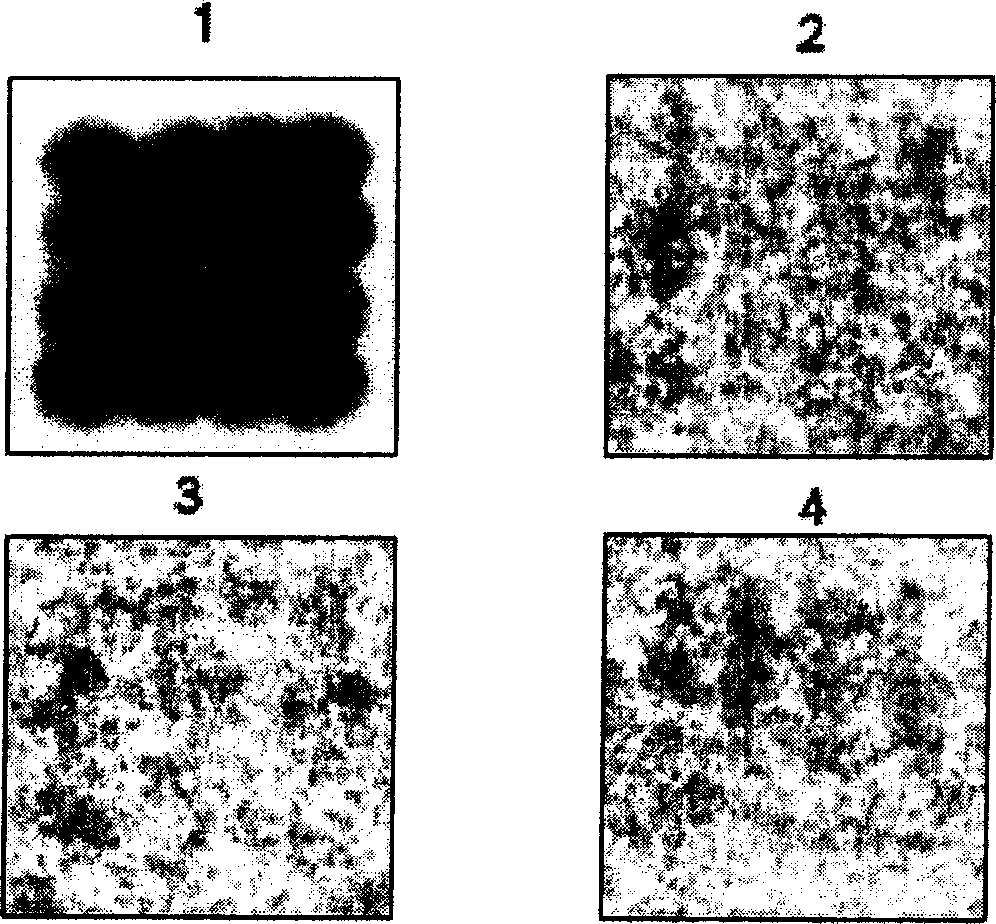
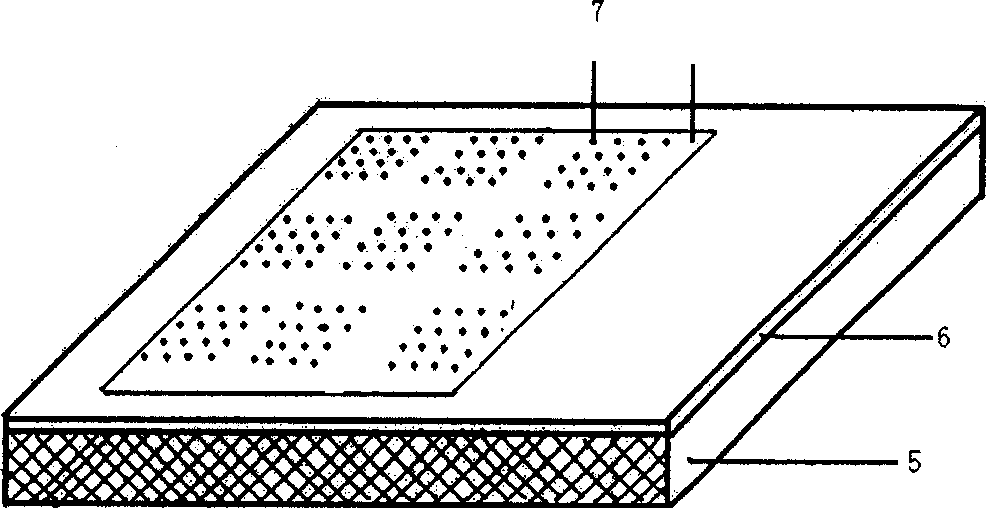
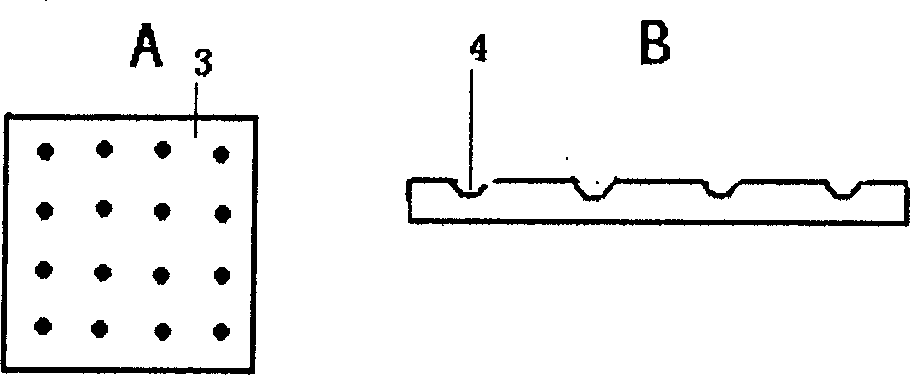
[0021] Such as figure 2 As shown, the gene chip is a film 6 covered with a positively charged material on the plane of the planar support carrier 5, and a dot matrix 7 is arranged on the surface of the film layer. Exogenous gene probes: 1) 35S promoter, 2) FMV35S promoter, 3) Nos terminator, 4) NPTII gene, 5) CryI (AC) gene, 6) Cry9c gene, 7) transformed CP4-EPSPS Gene, 8) 35s-CTP2, 9) CP4-EPSPS gene, 10) GOX gene, 11) Bar gene, 12) Barnase gene, 13) Barstar gene, 14) CryIA(a) gene, 15) Lectin gene, 16) Napin gene, 17) Zein gene, 18) Sad1 gene, 19) Lat52 gene or foreign gene fragment.
[0022] The dot matrix 7 is a dimple-like structure 8 arranged, and space intervals are maintained between the dot-matrix 7 , and the exogenous gene probes for transgenic plant judgment are respectively covered in each dimple-like structure 8 . The film layer 6 of positively charged material is a film layer o...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com