Reagent and method for detecting leucocythemia susceptibility
A kit and susceptibility technology, applied in biochemical equipment and methods, microbial assay/inspection, etc., can solve problems such as expression decline
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0029] Example 1. Obtaining SNP
[0030] Step 1: Extraction of genomic DNA
[0031] Using a small amount of peripheral blood leukocyte genomic DNA rapid extraction and purification kit (Shanghai Huashun Biological Engineering Co., Ltd.), 2ml of human whole blood was used to extract genomic DNA; after the concentration was adjusted to 50ng / μl, it was used for conventional PCR amplification.
[0032] Step 2: Obtain SNP
[0033] The genomic DNA extracted in step 1 is subjected to specific PCR amplification, and the amplified product is purified and then directly sequenced. The sequence of each PCR amplification product determined is compared to obtain the sequence difference and obtain the SNP. Among them, the PCR reaction conditions were 95°C for 3 minutes, 35 cycles of 95°C for 30 seconds, 68°C for 1 minute, and finally 72°C for 7 minutes. The primers are:
[0034] Sense primer: 5'-CGGGGAATCGGGCCTCTGC-3' (SEQ ID NO: 2)
[0035] Antisense primer: 5'-CAAGCTCCTCGTCCGCCATG-3' (SEQ ID N...
Embodiment 2
[0037] Example 2. Detection kit
[0038] Step 1: Extraction of DNA template
[0039] 2ml of human peripheral blood was drawn by conventional methods, and genomic DNA in these blood samples was extracted by conventional methods.
[0040] Step 2: PCR reaction
[0041] Prepare a detection kit for detecting the susceptibility of PDCD5 gene-related diseases, which contains the following primer pairs that can amplify 154 and 170 SNPs:
[0042] Sense primer: 5'-GCTCCGGGCTGGATTGGTG-3' (SEQ ID NO: 6)
[0043] Antisense primer: 5'-CATGGCTCGGCGTCAGCG-3' (SEQ ID NO: 12)
[0044] The PCR reaction conditions were 95°C, 3 minutes, 35 cycles of 95°C for 30 seconds, 68°C for 1 minute, and finally 72°C for 7 minutes.
[0045] Step 3: Genetic analysis
[0046] The amplified product was genotyped with RFLP technology. Because the 170th G polymorphism produced a Nar I endonuclease site and the 154th A destroyed a Nar I restriction site, the Nar I enzyme was used for the analysis. Enzyme digestion can ...
Embodiment 3
[0047] Example 3. Analysis of the functional effects of the 154A>G / 170G>A polymorphism in the PDCD5 promoter region Step 1. Construction of the reporter vector for the 154G / 170A genotype and the promoter region of the 154A / 170G genotype
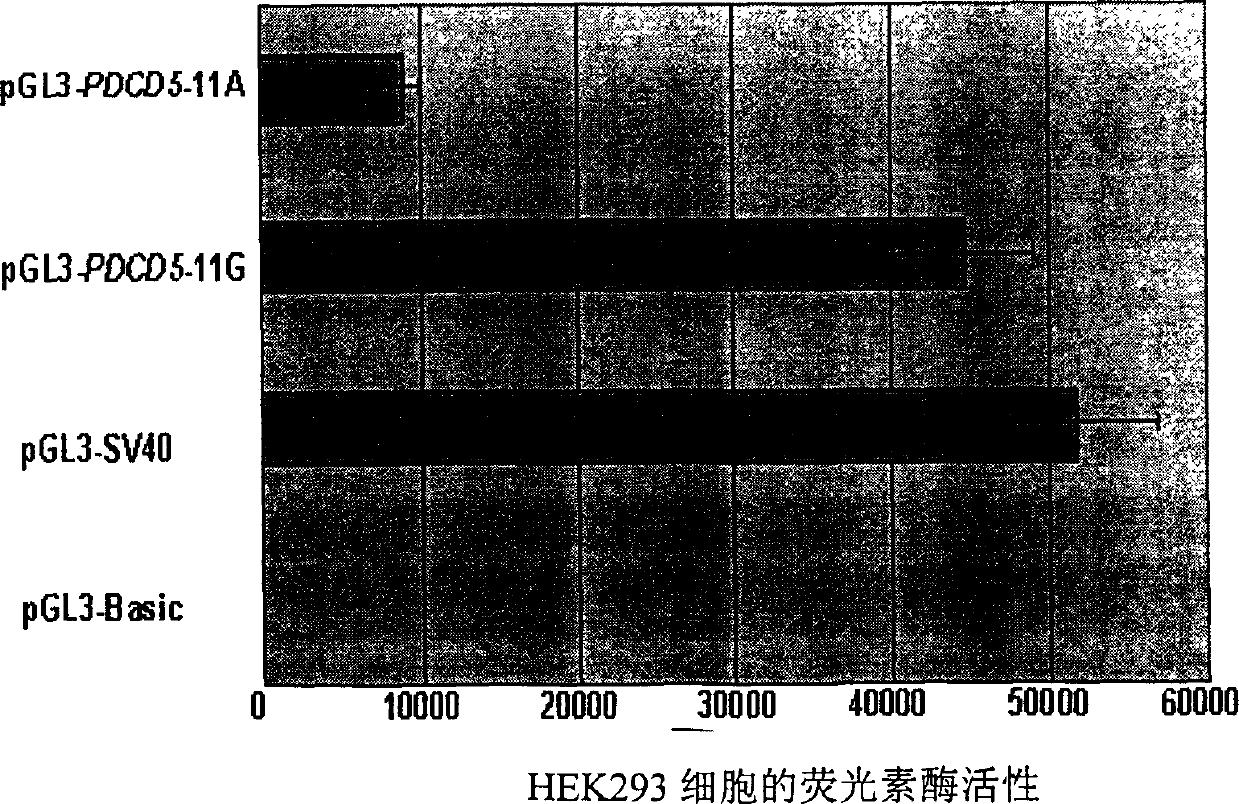
[0048] A 743 bp DNA fragment containing the 154G / 170A mutant genotype and the 154A / 170G wild genotype of the PDCD5 promoter region from the 643th position upstream of the transcription start site to the 91 position downstream of the transcription start site was cloned into pGL3-Basic( Promega company) report plasmid, construct the luciferase reporter gene plasmid pGL3-PDCD5-170A (154G / 170A) containing 154G / 170A mutant type and pGL3-PDCD5-170G (154A / 170G) containing 154A / 170G wild type pGL3-PDCD5-170G (154A / 170G) Luciferase reporter gene plasmid.
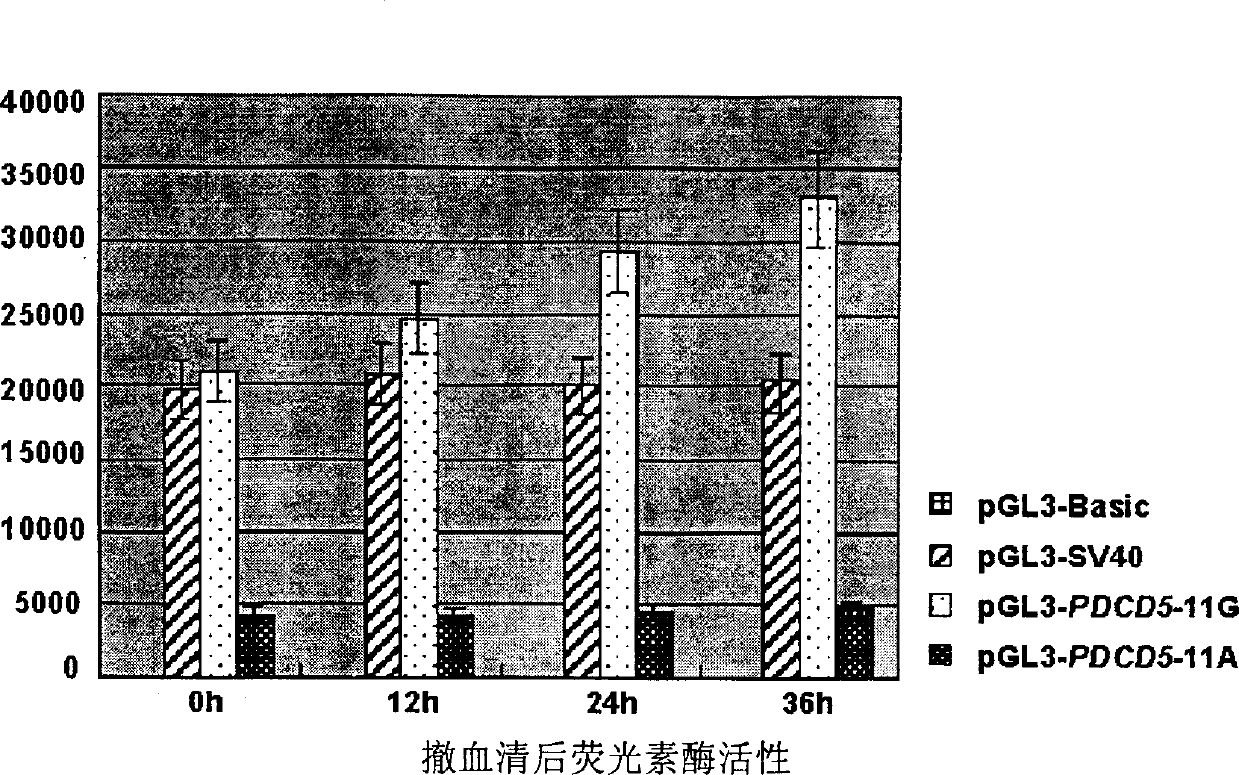
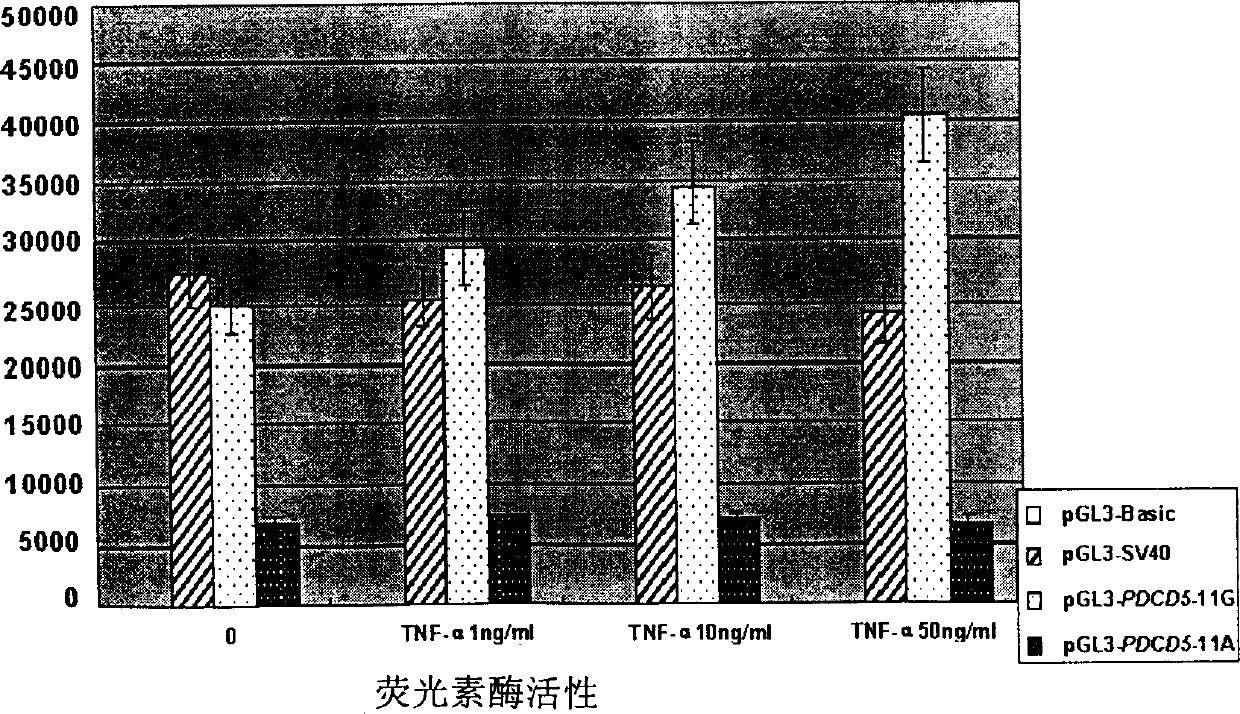
[0049] Step 2. Detection of promoter activity of 154G / 170A genotype and 154A / 170G genotype
[0050] The negative control pGL3-Basic, the positive control pGL3-SV40 (Promega), pGL3-PDCD5-170A and pGL3-PD...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com