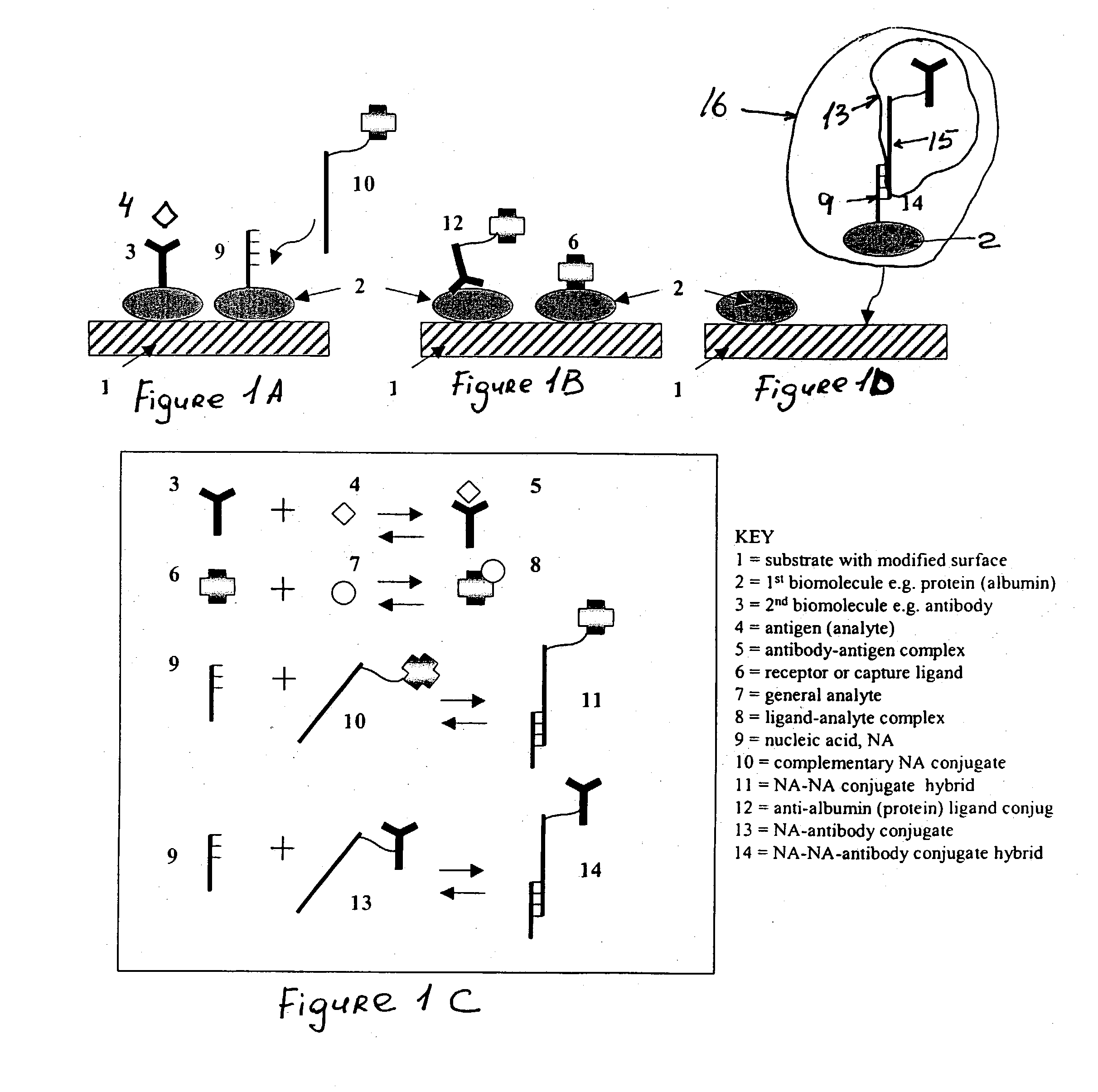
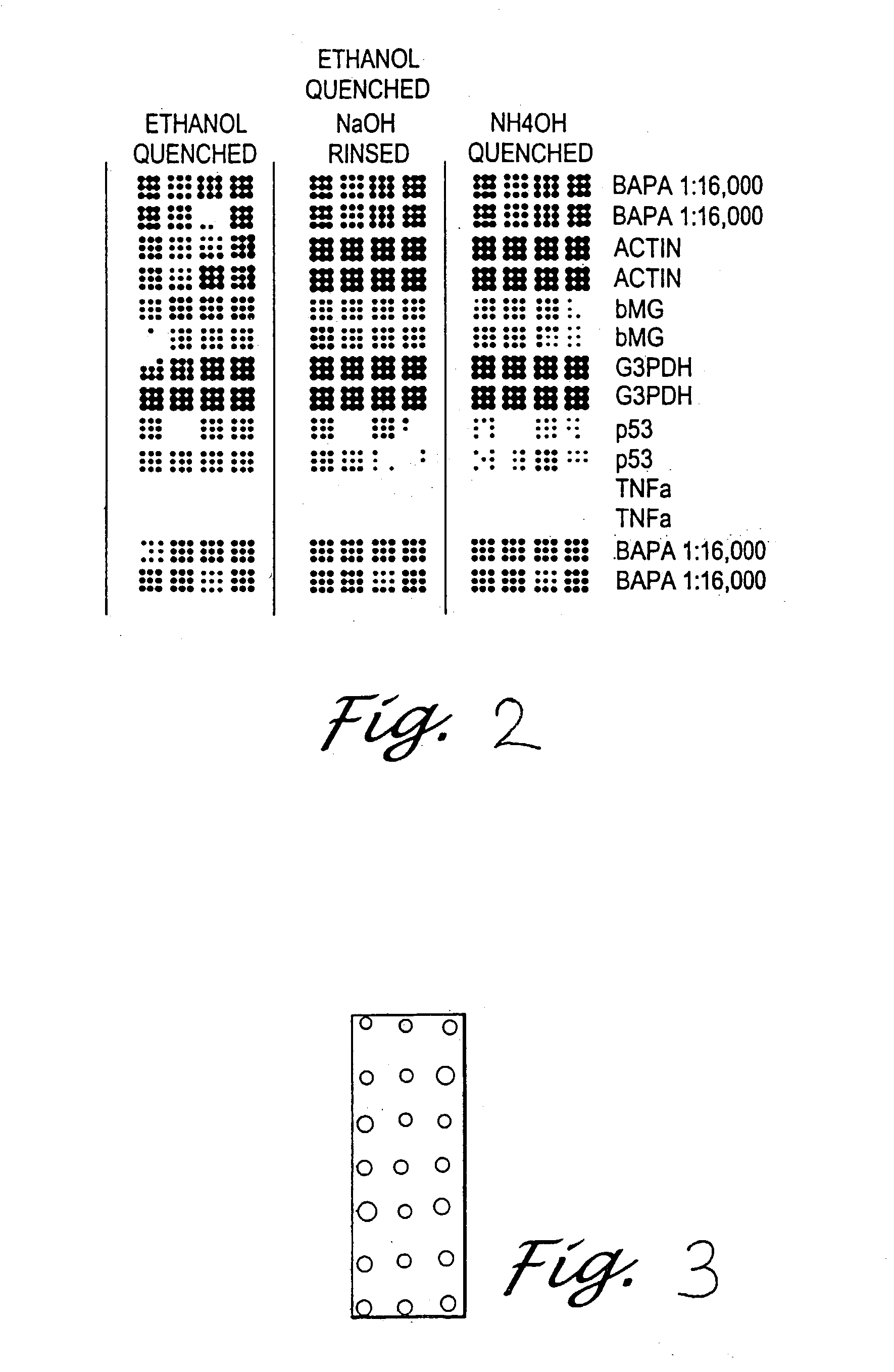
Immobilization of biomolecules on substrates by attaching them to adsorbed bridging biomolecules
a biomolecule and substrate technology, applied in the field of immobilization of biomolecules on substrates, can solve the problems of difficult to obtain accurate and reproducible data with arrays formed by direct synthesis, undesirable non-specific electrostatic binding of nucleic acids, and difficulty in obtaining direct synthesis accurate and reproducible data, etc., to achieve cost-effective production, reduce production costs, and simplify the task of making arrays
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
example 2
[0097] Preparation of Protein Array
[0098] Human IgG attachment: Polypropylene film was surface aminated by a radio-frequency discharge as described in Example 1. Diluted Human IgG (Pierce Chemicals, cat. # 31877, 28 mg / mL) stock to 1 mg / mL in sodium bicarbonate (50 mM, pH 9) and 4% sodium sulfate. Twenty-one 0.5 .mu.L spots were pipetted onto an amino polypropylene strip 2 cm wide and 8 cm long. Attachment reaction was allowed to proceed for 60 min. at 25.degree. C. The film was rinsed with Casein solution (1 mg / mL in 50 mM sodium carbonate, 0.1 5M NaCl pH 9) for 60 min. at 25.degree. C., and then rinsed twice in deionized water and by 1.times. TBS, 0.02% Tween-20, pH 7.4 briefly. The strip was then used for binding assay.
[0099] Conjugation with goat anti-human IgG alkaline phosphatase: The 200 .mu.L of diluted goat anti-human IgG alkaline phosphatase (Pierce Chemicals, cat. # 31310) solution 1:1000 in blocking buffer (1.times.TBS, 1 mg / mL Casein, 0.02% Tween-20 pH 7.4) was pipetted...
PUM
| Property | Measurement | Unit |
|---|---|---|
| temperatures | aaaaa | aaaaa |
| pH | aaaaa | aaaaa |
| temperatures | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com