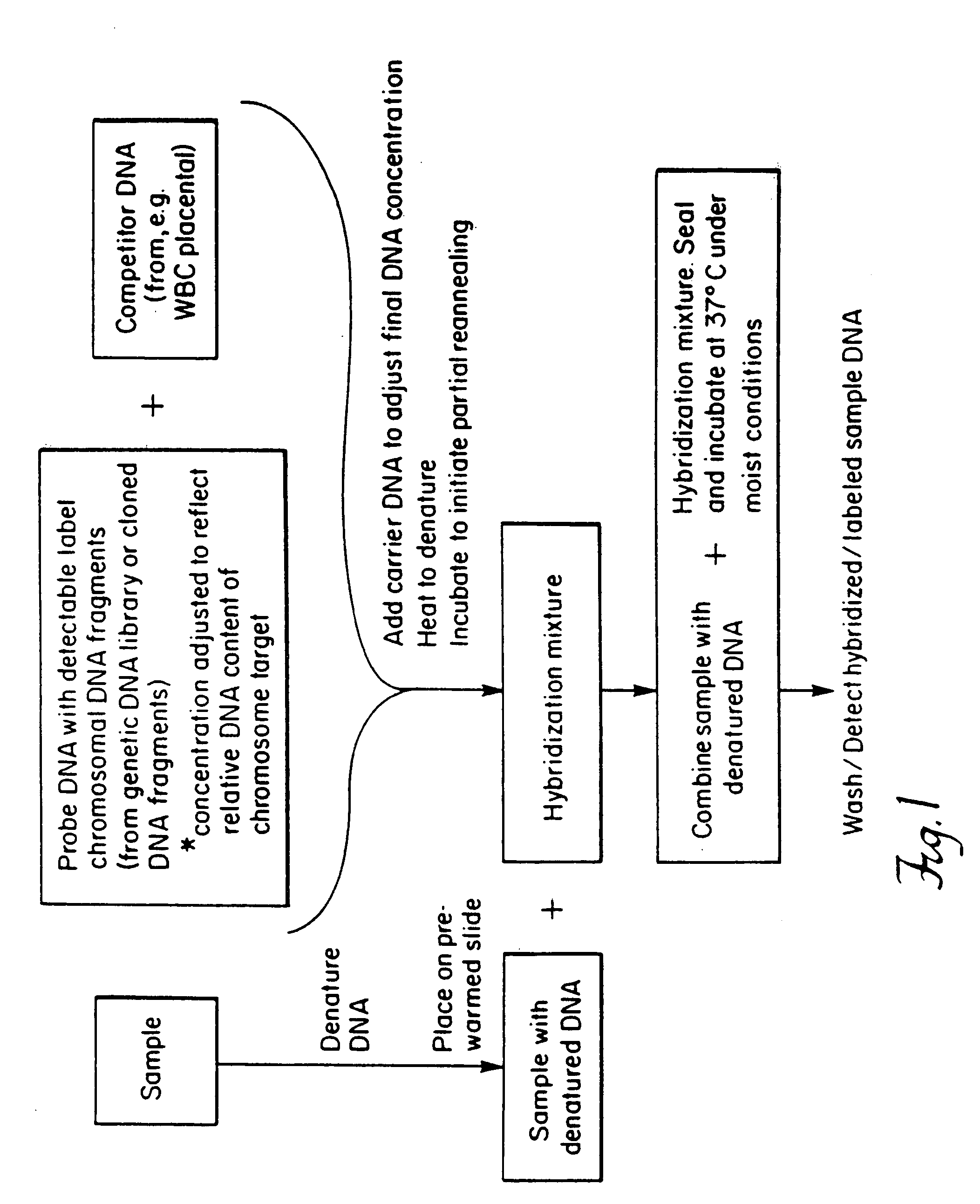
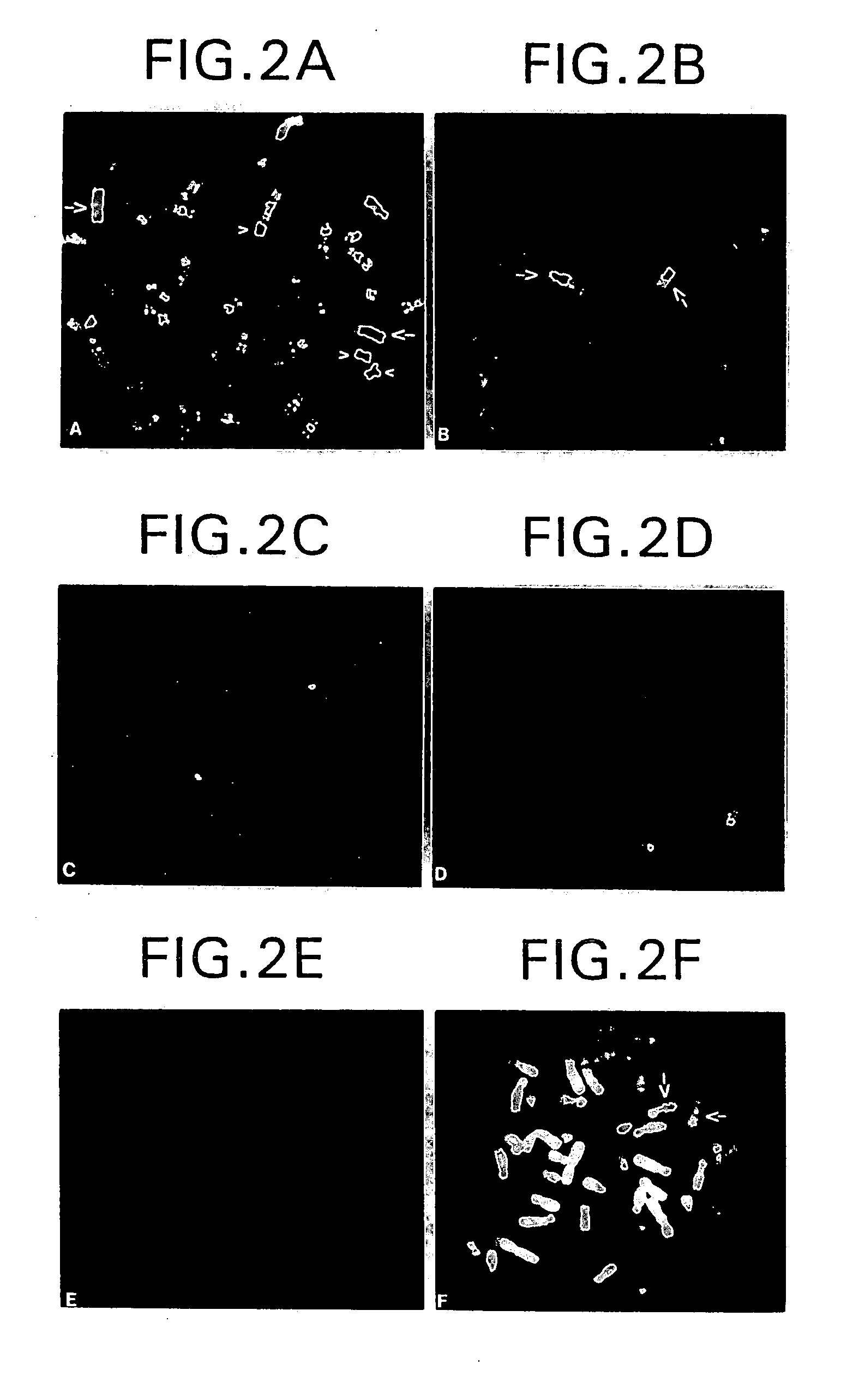
Delineation of individual human chromosomes in metaphase and interphase cells by in situ suppression hybridization
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
example 1
Cyto-Specific Staining of Individual Human Chromosomes Using Genomic DNA Libraries in CISS Hybridization
DNA Libraries
[0120] The following human chromosome genomic libraries were obtained from the American Type Culture Collection: LAO1NSO1 (chromosome 1), LL04NSO1 (chromosome 4), LA07NsO1 (chromosome 7), LL08NS02 (chromosome 8), LA13NS03 (chromosome 13), LL14NSO1 (chromosome 14), LL19NSO1 (chromosome 18), LL20NSO1 (chromosome 20), LL21NS02 (chromosome 21), LA22NS03 (chromosome 22), LAOXNLO1 (chromosome X). Amplification of these phage libraries on agar plates (using LE 392 cells as the bacterial host), purification of the X phages and extraction of phage-DNA pools were carried out according to standard protocols. Maniatis, T. et al., Molecular Cloning: A Laboratory Manual, Cold Spring Harbor Laborabory, Cold Spring Harbor, N.J. (1982).
Preparation of Metaphase Spreads and Fibroblast Cells
[0121] Phytohemagglutinin-stimulated lymphocytes from a normal adult male (46, XY) were cult...
example 2
Detection of Chromosome Aberrations in Tumor Cells by CISS Hybridization Using Chromosome-Specific Library Probes
Cells
[0145] TC 593 is a pseudotetraploid cell line (modal chromosome number, 83) established from a human glioblastoma; it grows in a flat, spreading fashion and contains many process. TC 620 is pseudotriploid with a modal chromosome number of 64 and was established from a human oligodendroglioma; it grows in an epithelial fashion. Both cell lines have been described in detail. Manuelidis, L. and E. E. Manuelidis, In: Progress in Neuropathology, Vol. 4, 235-266, Raven Press, N.Y. (1979). The present experiments made use of subclones C2B (TC 593) and C2B (TC 620) at approximately 180 passages after repeated subcloning from a single cell of the original tumor line cultured as previously described by Manuelidis and Manuelidis (see reference above). Standard hypotonic treatment and acid / methanol fixation of the cells were employed. Cremer et al., Exp. Cell Res., 176:199-22...
example 3
Rapid Detection of Human Chromosome 21 Aberrations By In Situ Hybridization
DNA Probes
[0174] All plasmids contain inserts of human chromosome 21 that were mapped to 21q22.3. Moisan, J. P., Mattei, M. G., Baeteman-Volkel, M. A., Mattei, J. F., Brown, A. M. C., Garnier, J. M., Jeltsch, J. M., Masiakowsky, P., Roberts, M. & Mandel, J. L. (1985) Cytogenet. Cell Genet. 40, 701-702 (abstr.). Tanzi, R., Watkins, P., Gibons, K., Faryniarz, A., Wallace, M., Hallewell, R., Conneally, P. M. & Gusella, J. (1985) Cytogenet. Cell Genet. 40, 760 (abstr.). Van Keuren, M. L., Watkins, P. C., Drabkin, H. A., Jabs, E. W., Gusella, J. F. & Patterson, D. (1986) Am. J. Hum. Genet. 38, 793-804. Nakai, H., Byers, M. G., Watkins, P. C., Watkins, P. A. & Shows, T. B. (1987) Cytogenet. Cell Genet. 46, 667 (abstr.). Munke, M., Foellmer, B., Watkins, P. C., Cowan, J. M., Carroll, A. J., Gusella, J. F. & Fracke, U. (1988) Am. J. Humm. Genet. 42, 542-549. All inserts were either known or verified by Southern bl...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com