Meningococcus adhesins nada, app and orf 40
a technology of meningococcus and adhesins, applied in the field of biochemistry, can solve the problem that the pathogen does not reveal everything, and achieve the effect of enhancing the biological activity of a particular region and minimizing misfolding
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
y
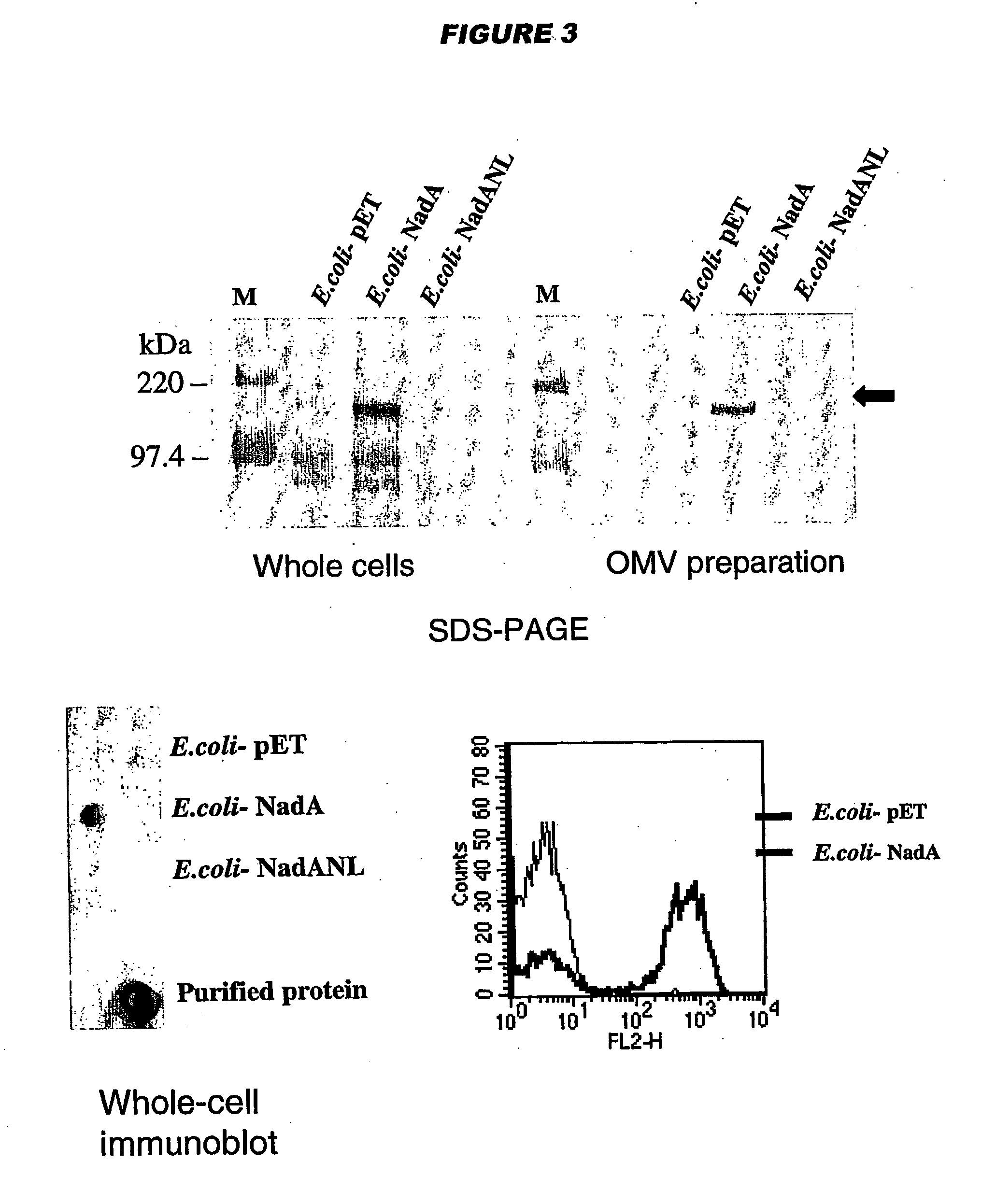
[0198] NadA shows homology to (a) YadA of enteropathogenic Yersinia, a non-pilus associated adhesin implicated in virulence [Cornelis (1998) Microbiol. Mol. Biol. Rev. 62:1315-1352.] and (b) UspA2 of Moraxella catarrhalis, a protein involved in serum resistance and a protective antigen [Chen et al. (1999) Infect. Immun. 67:1310-1316.]. Sequence similarity is mainly clustered in the carboxyl terminal region (56-63% identity in the last 70 amino acids). Outside this region the level of identity drops to 23-25%.
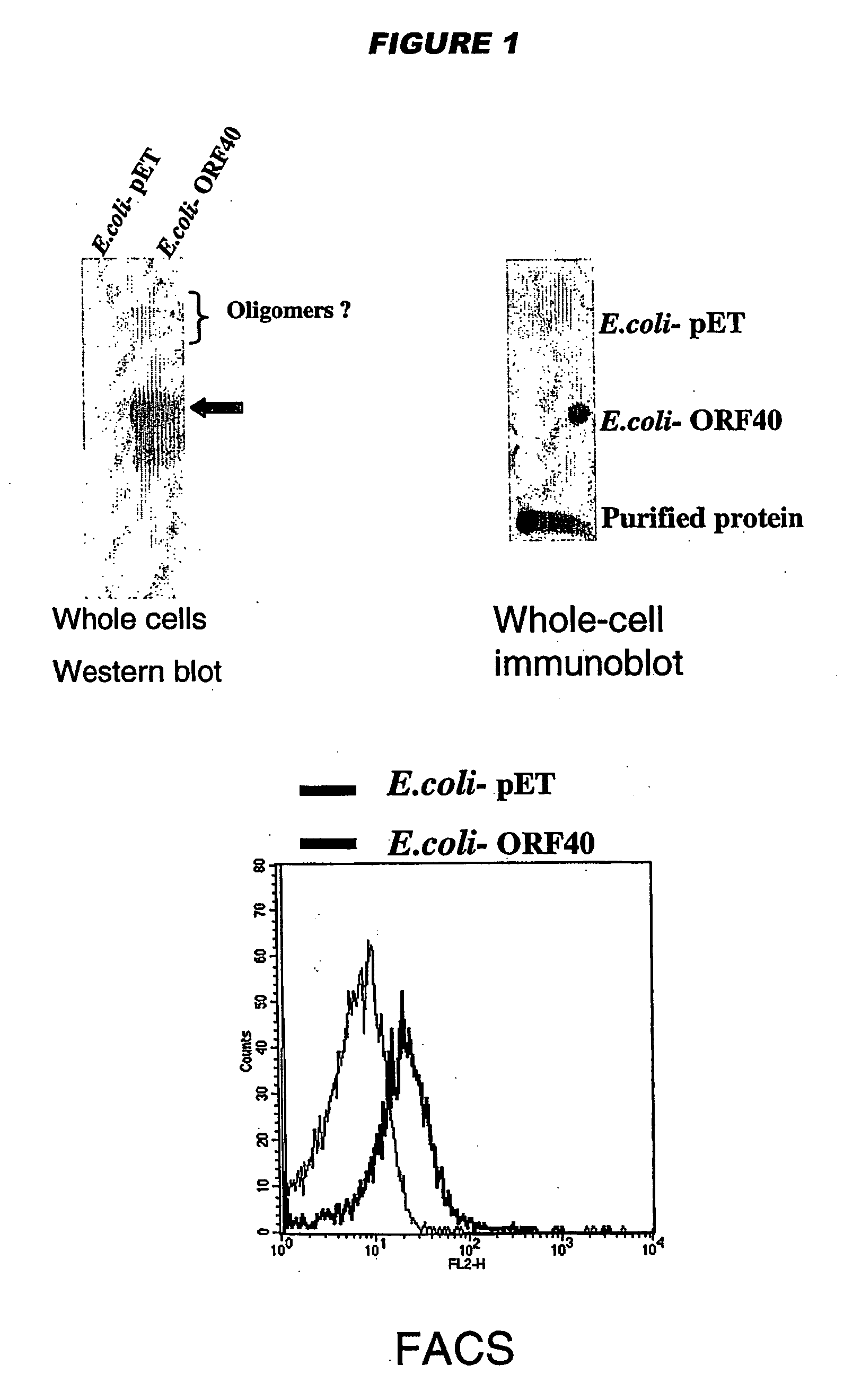
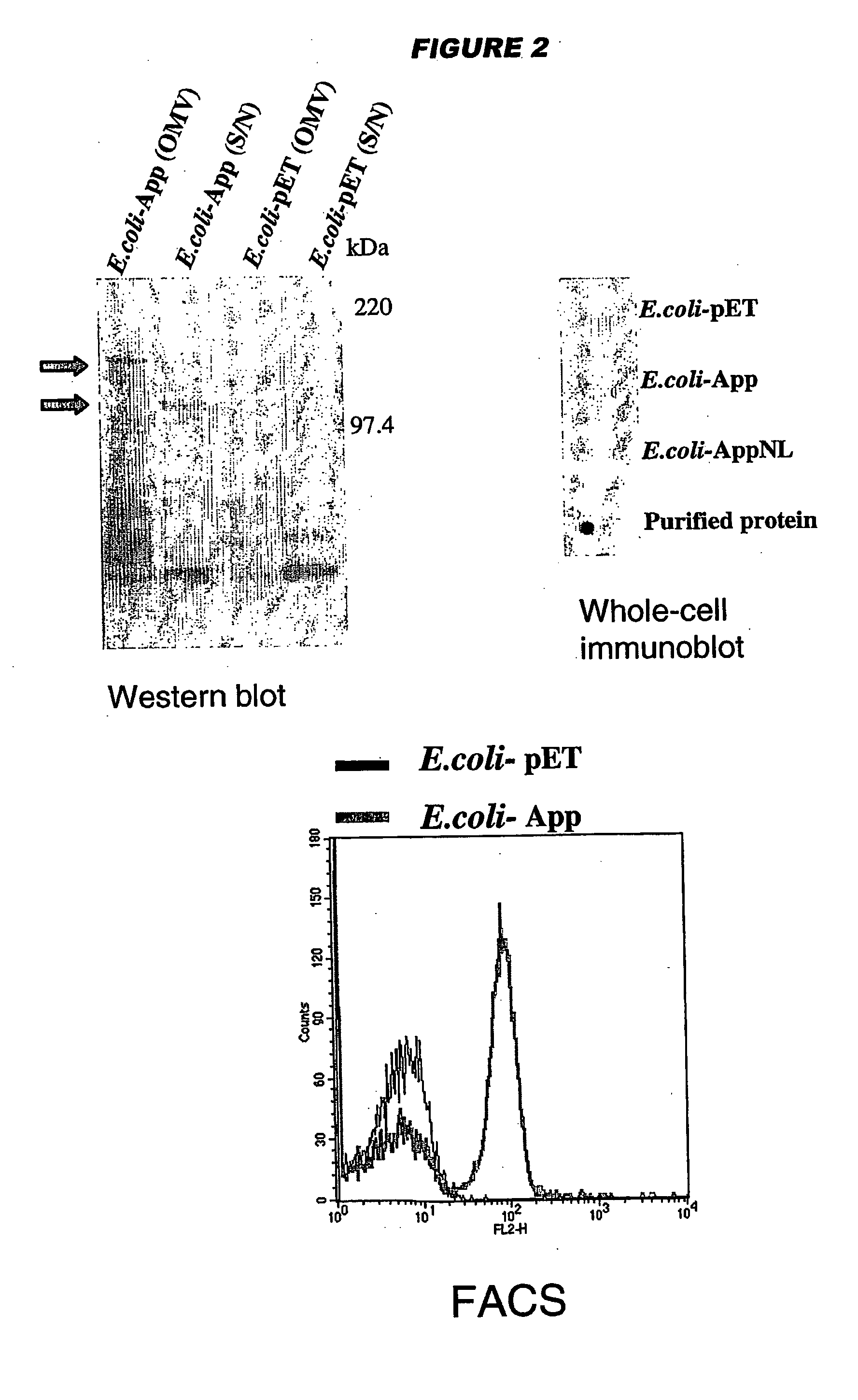
[0199] YadA and UspA2 have been identified as adhesins [Hoiczyk et al. (2000) EMBO J. 19:5989-5999]. Both proteins form very stable and difficult-to-dissociate high molecular weight oligomers (150-200 kDa) anchored to the outer membrane. NadA has also been found to form very stable high molecular weight aggregates on the outer membrane of meningococcus.
[0200] The amino acid sequence of NadA was analysed [Nielsen et al. (1997) Protein Engineering 10:1-6; Levin & Garner (1988) ...
PUM
| Property | Measurement | Unit |
|---|---|---|
| pH | aaaaa | aaaaa |
| pH | aaaaa | aaaaa |
| pH | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com