Method for automated, large-scale measurement of the molecular flux rates of the proteome or the organeome using mass spectrometry
a technology of proteome and organeome, which is applied in the field of large-scale measurement of the molecular flux rate of the proteome or the organeome using mass spectrometry, can solve the problems of not providing information about biochemical function (phenotype) in a living system, gene expression chips do not solve the central problem of phenotype and function in biochemistry, and the log-jam arises
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
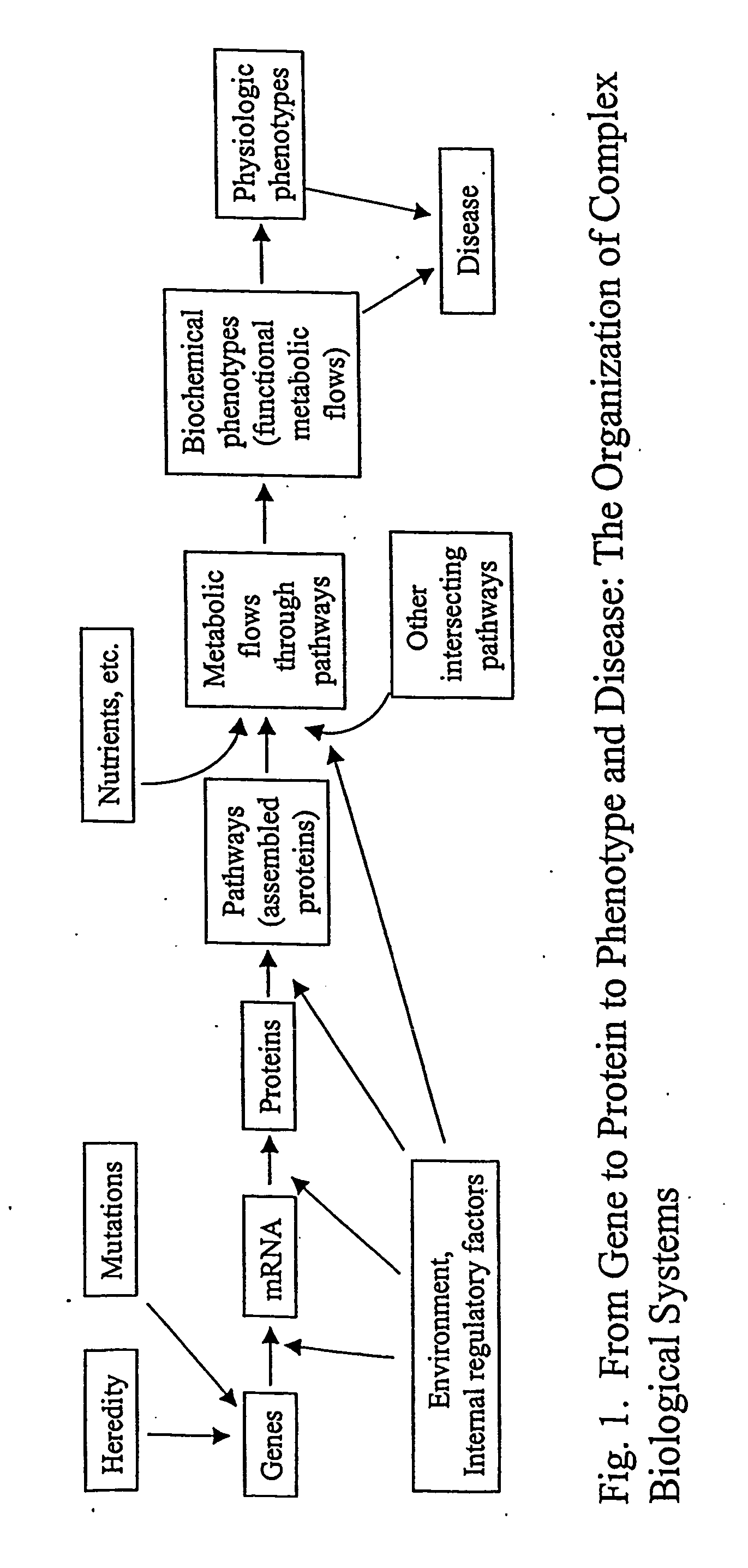
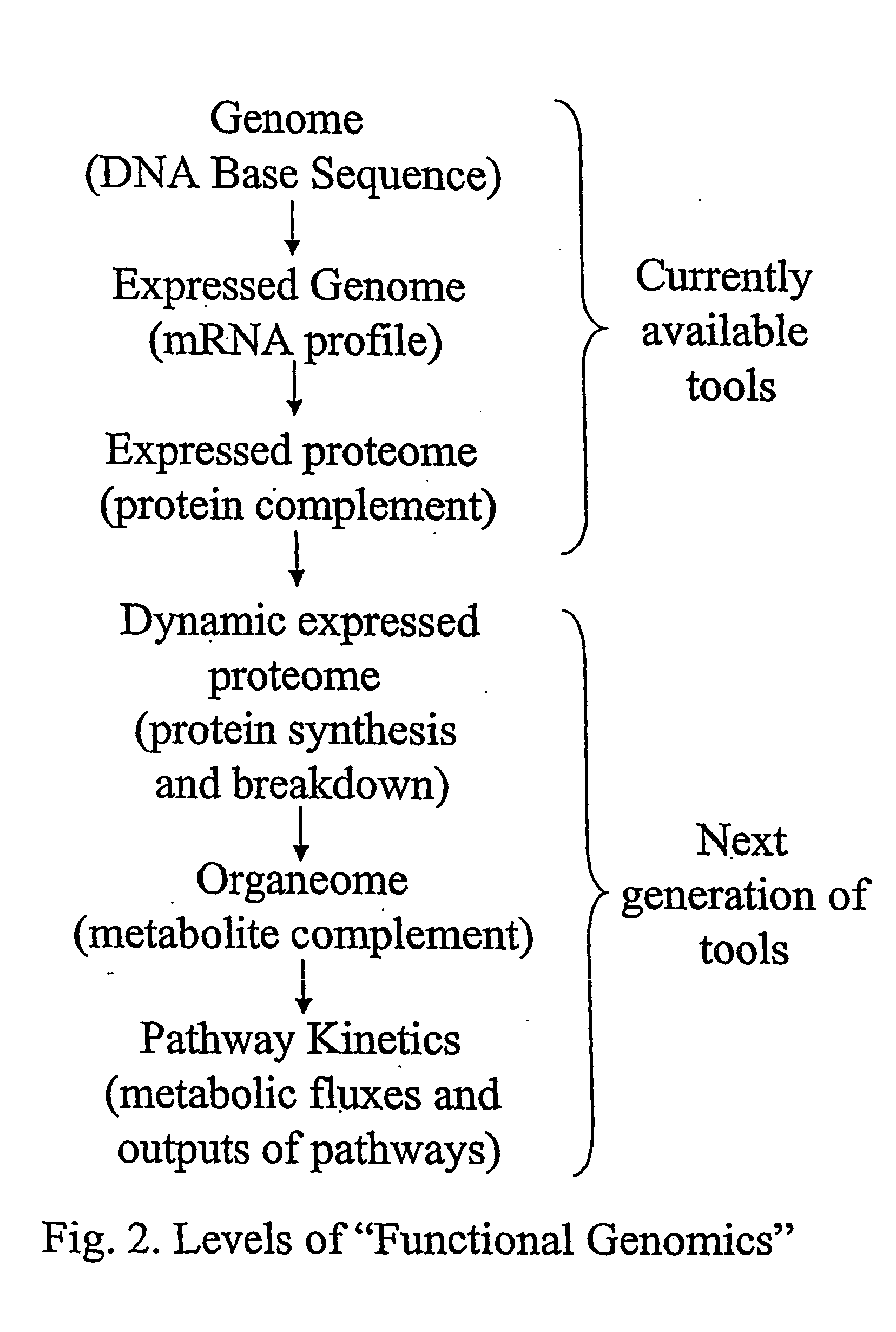
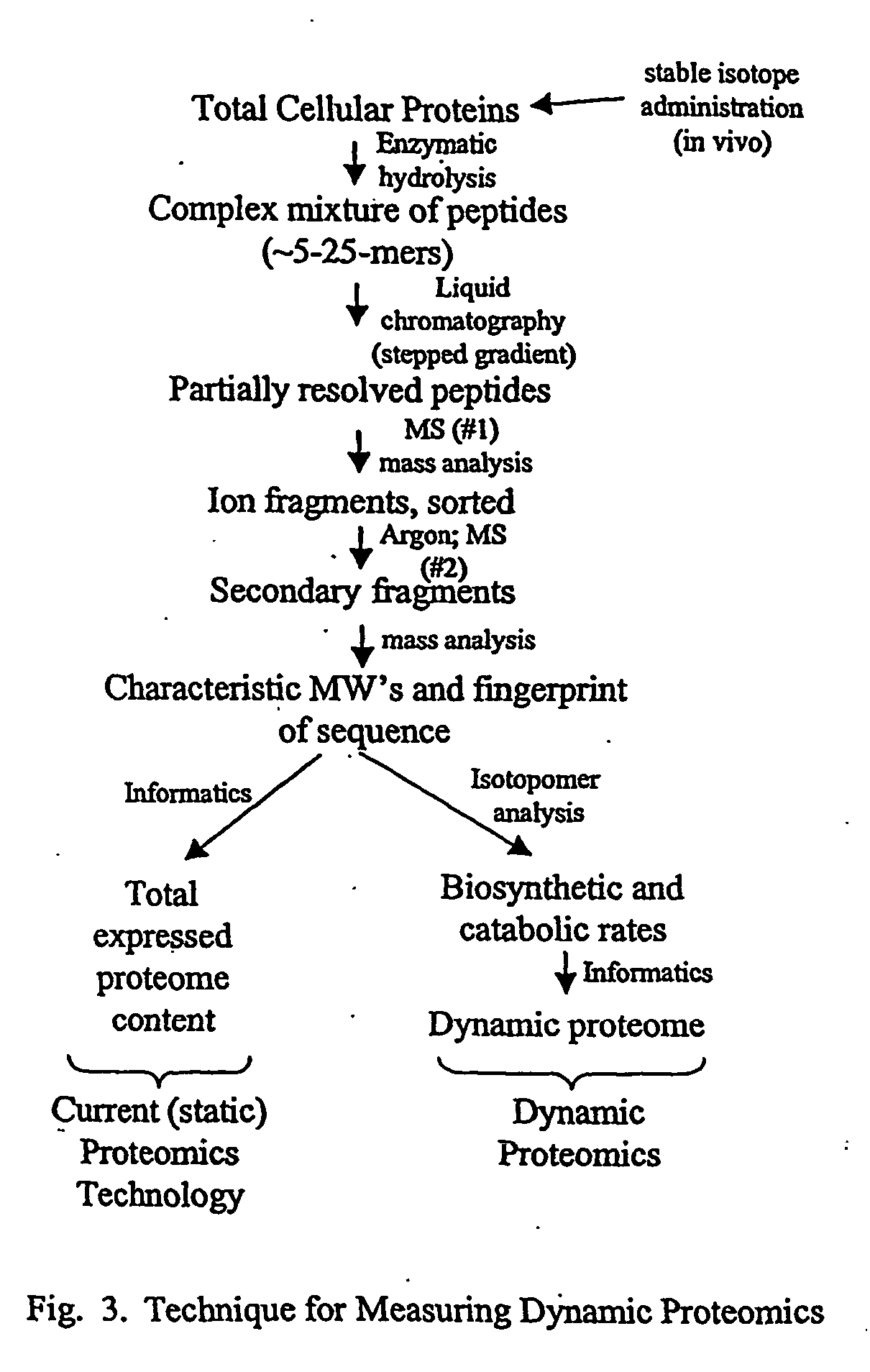
[0029] The inventor has discovered a method of determining molecular flux rates (i.e., synthesis, breakdown and removal rates) of a plurality of proteins or a plurality of organic metabolites. First, an isotope-labeled precursor molecule is administered to a cell, tissue, or organism. The molecular flux rates of a plurality of proteins or a plurality of organic metabolites are then determined.
I. General Techniques
[0030] The practice of the present invention will employ, unless otherwise indicated, conventional techniques of molecular biology (including recombinant techniques), microbiology, cell biology, biochemistry and immunology, which are within the skill of the art. Such techniques are explained fully in the literature, such as, Molecular Cloning: A Laboratory Manual, second edition (Sambrook et al., 1989) Cold Spring Harbor Press; Oligonucleotide Synthesis (M. J. Gait, ed., 1984); Methods in Molecular Biology, Humana Press; Cell Biology: A Laboratory Notebook (J. E. Cellis,...
PUM
| Property | Measurement | Unit |
|---|---|---|
| Force | aaaaa | aaaaa |
| Electrical conductance | aaaaa | aaaaa |
| Electrical conductance | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com