Process for bacterial production of polypeptides
a polypeptide and bacterial cell technology, applied in the direction of peptides, hormone peptides, peptides/protein ingredients, etc., can solve the problems of inability to translate easily and efficiently, inability to conventionally isolate heterologous polypeptides from gram-negative bacteria, and inability to recombinant protein products. to achieve the effect of reducing the co-recovery of cellular debris, high efficiency and high efficiency
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
example i
[0099] IGF-I and T4-Lysozyme Nucleic Acid Co-expression Background
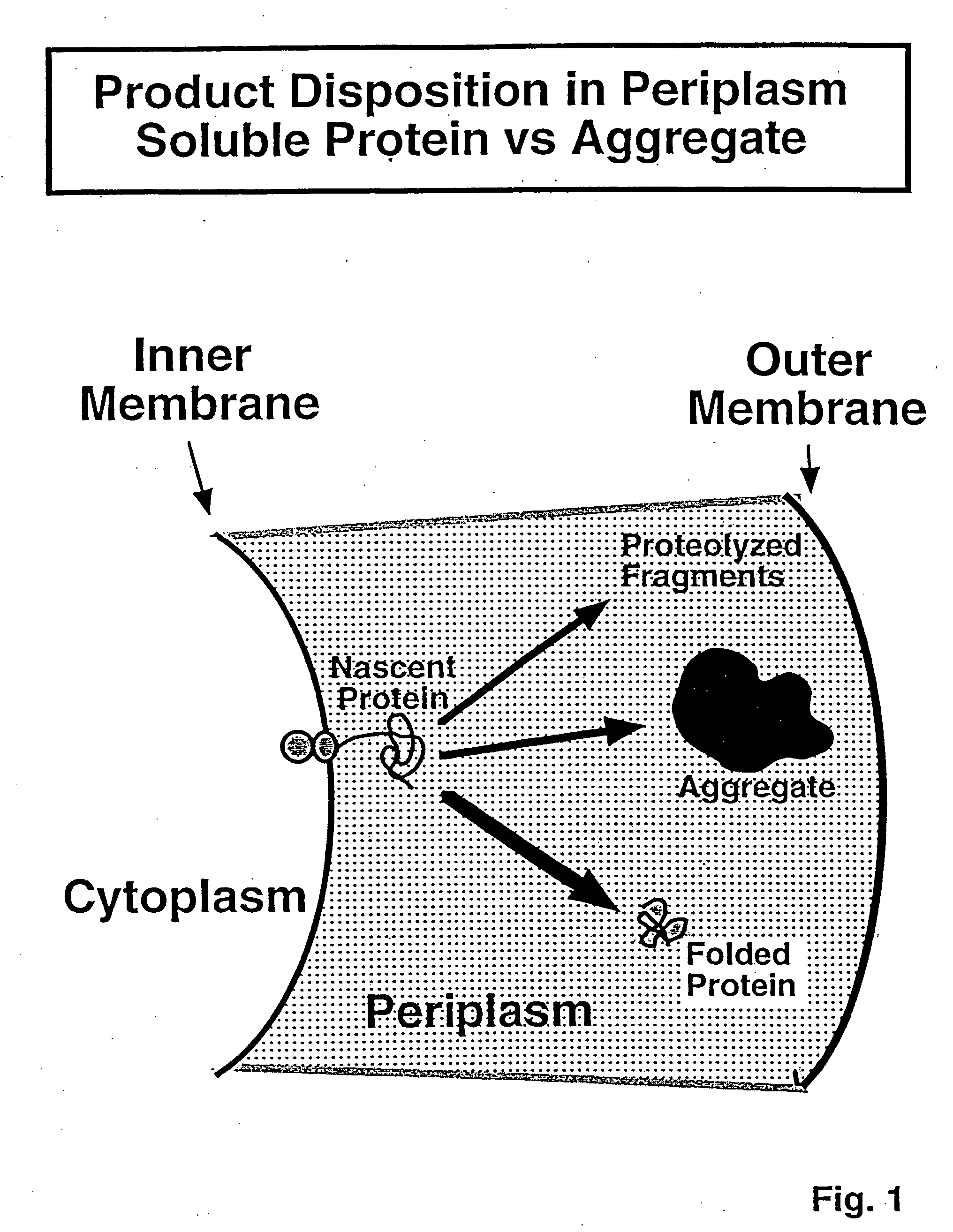
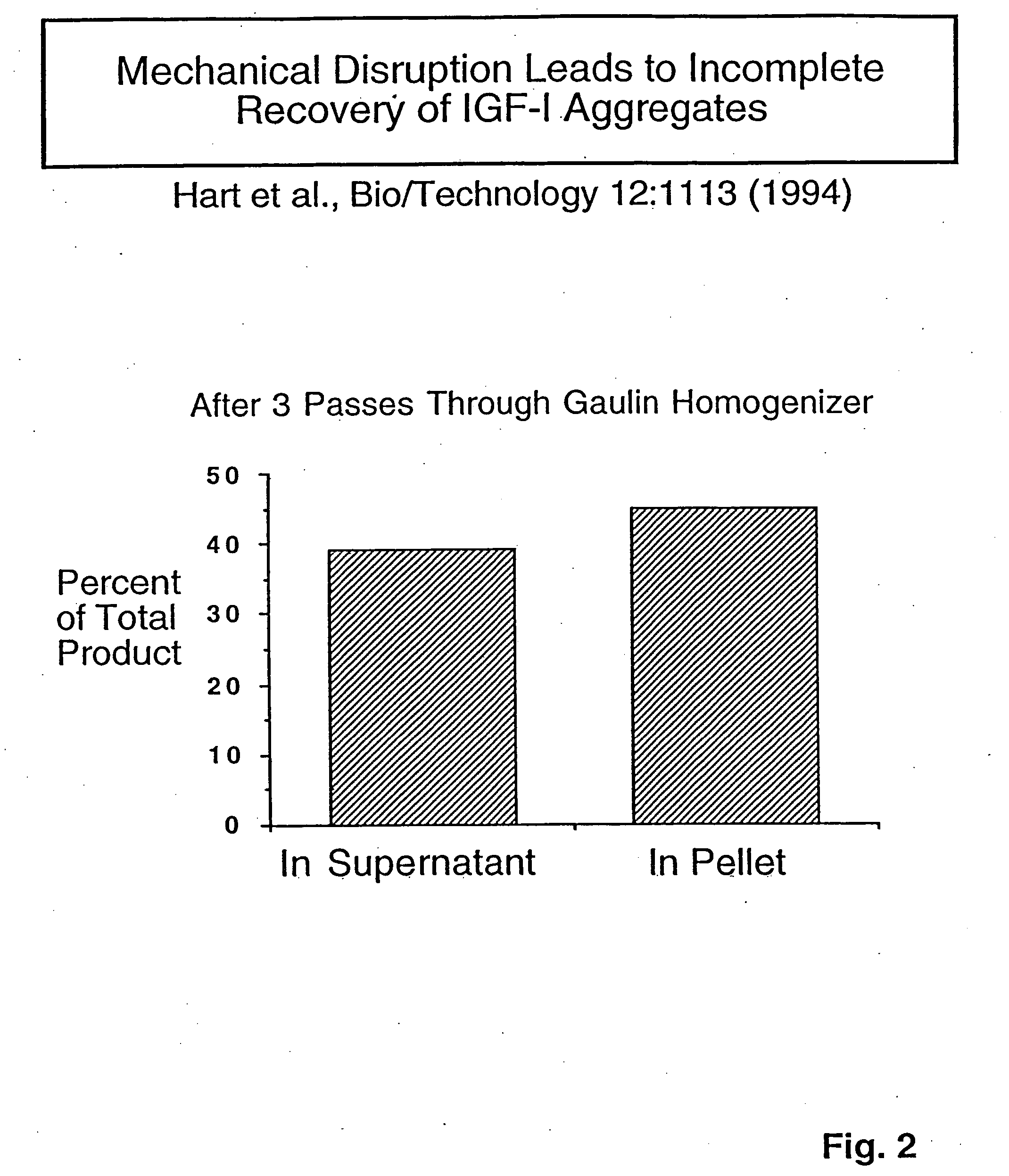
[0100] IGF-I was selected as a first protein for evaluation of refractile particle recovery due to large-scale needs. For this evaluation, a strategy was mapped out involving genetic manipulation of the host organisms to improve the release of the retractile particles from cell-wall structures.
Materials & Methods:
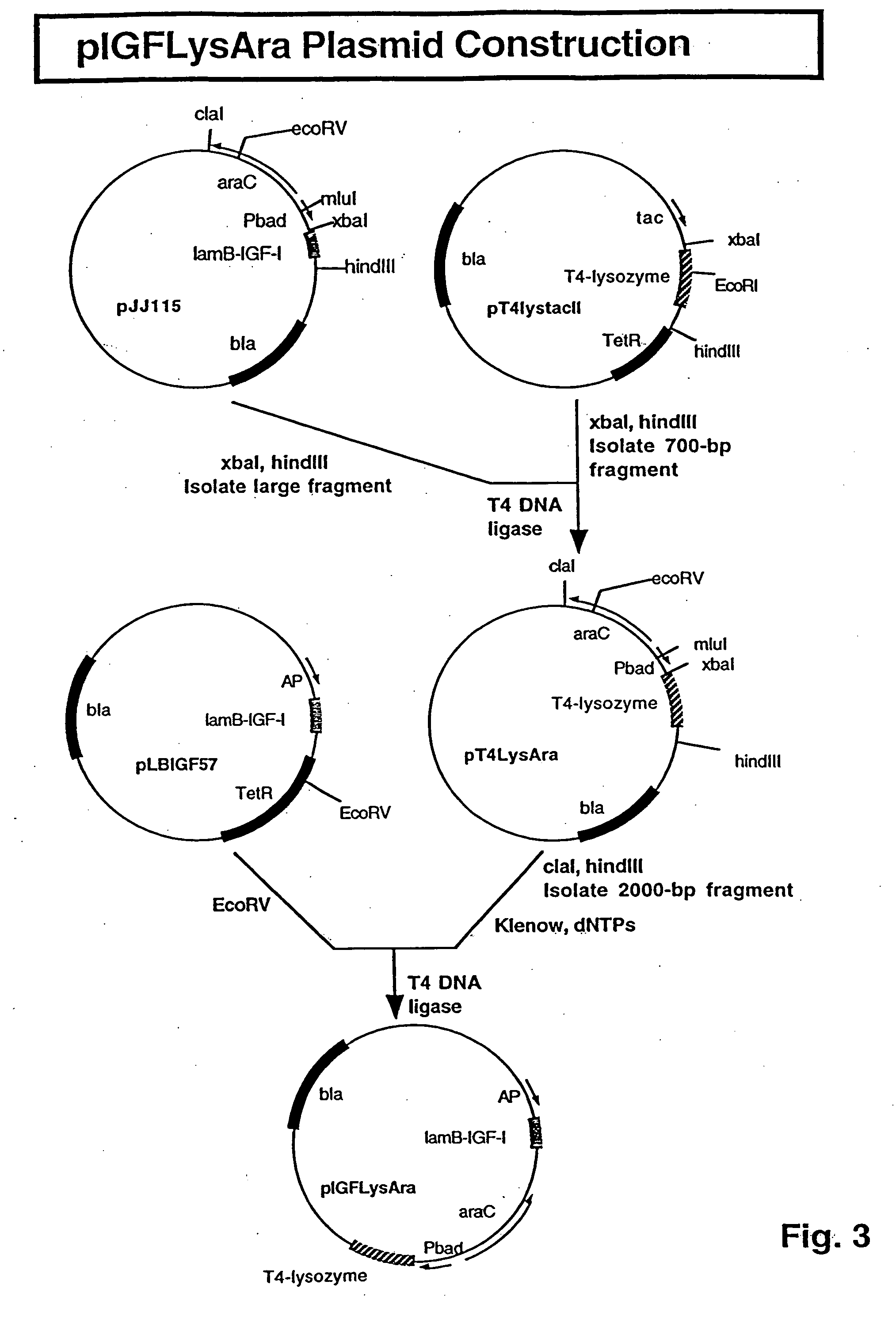
[0101] pIGFLysAra Plasmid Construction: In pIGFLysAra, the IGF-I encoding sequence has a lamB signal sequence for secretion into the periplasm, and was placed behind the alkaline phosphatase promoter (AP). The T4-lysozyme gene was placed behind the ara promoter for cytoplasmic accumulation of the gene product.
[0102] Details of the construction of the original plasmid, pT4lystacII, have been described in Gene, 38: 259-264 (1985). Intermediate plasmids were made to move the T4-lysozyme gene behind the ara promoter. Subsequently, the ara promoter-T4-lysozyme gene cassette was inserted into the IGF-I plasmid...
example ii
[0142] VEGF or DNase and T4-Lysozyme Nucleic Acid Co-Expression Background
[0143] It was important to determine if the T4-lysozyme nucleic acid co-expression technology had general application across other processes involving refractile particles. E.-coli-produced VEGF (a 21 kD protein) and DNase (a 31.9-kilodalton protein) were two additional products known to accumulate in the periplasmic space as refractile particles and therefore suitable proteins for evaluation. It was difficult to predict if T4-lysozyme nucleic acid co-expression would bring similar benefits to product recovery since it was not known if the physical properties of the refractile particles of VEGF and DNase differ significantly from that of the IGF-I refractile particles.
[0144] For efficient evaluation of the T4-lysozyme nucleic acid co-expression approach in multiple processes, a separate plasmid for the expression of nucleic acid encoding T4-lysozyme, pJJ153, was constructed. It was used in the co-transformat...
PUM
| Property | Measurement | Unit |
|---|---|---|
| temperature | aaaaa | aaaaa |
| temperature | aaaaa | aaaaa |
| molecular weight | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com