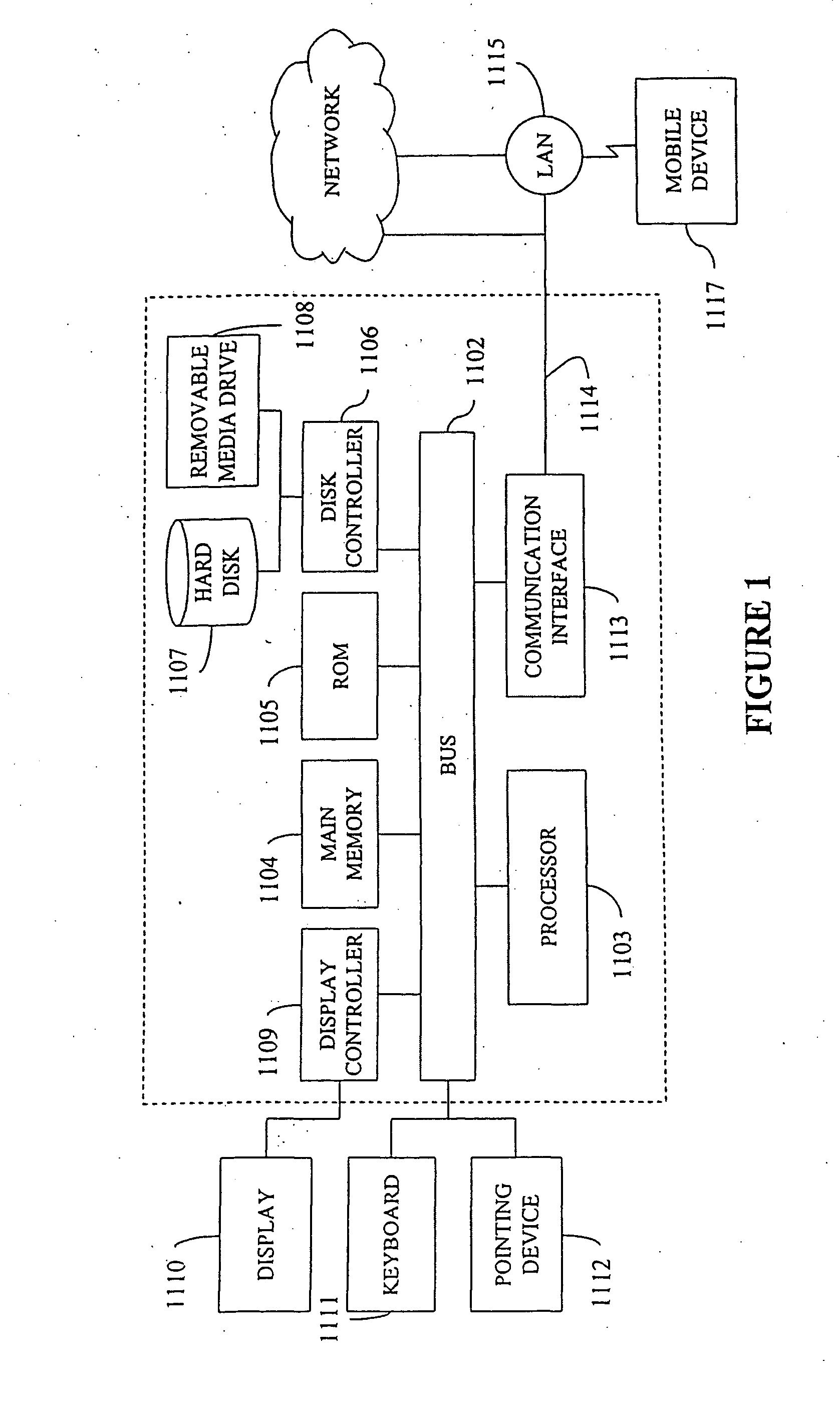
System, method, device, and computer program product for extraction, gathering, manipulation, and analysis of peak data from an automated sequencer
a technology of automated sequencers and computer programs, applied in the field of system, method, device and computer program products, can solve the problems of not having a tool available to allow the extensive and efficient retrieval of raw data, and achieve the effect of high throughput analysis of data
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
example 1.1
Mice and Parasites
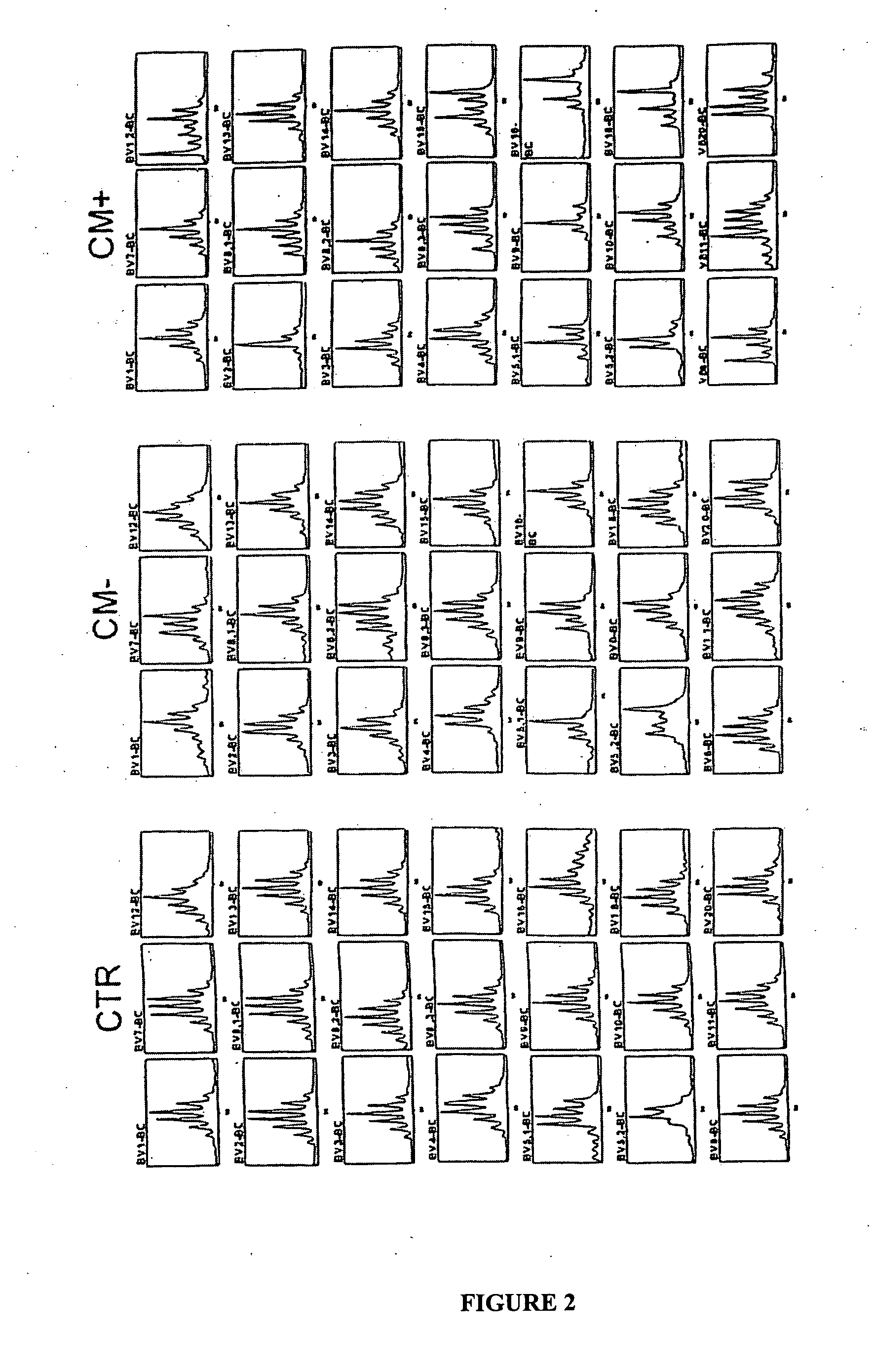
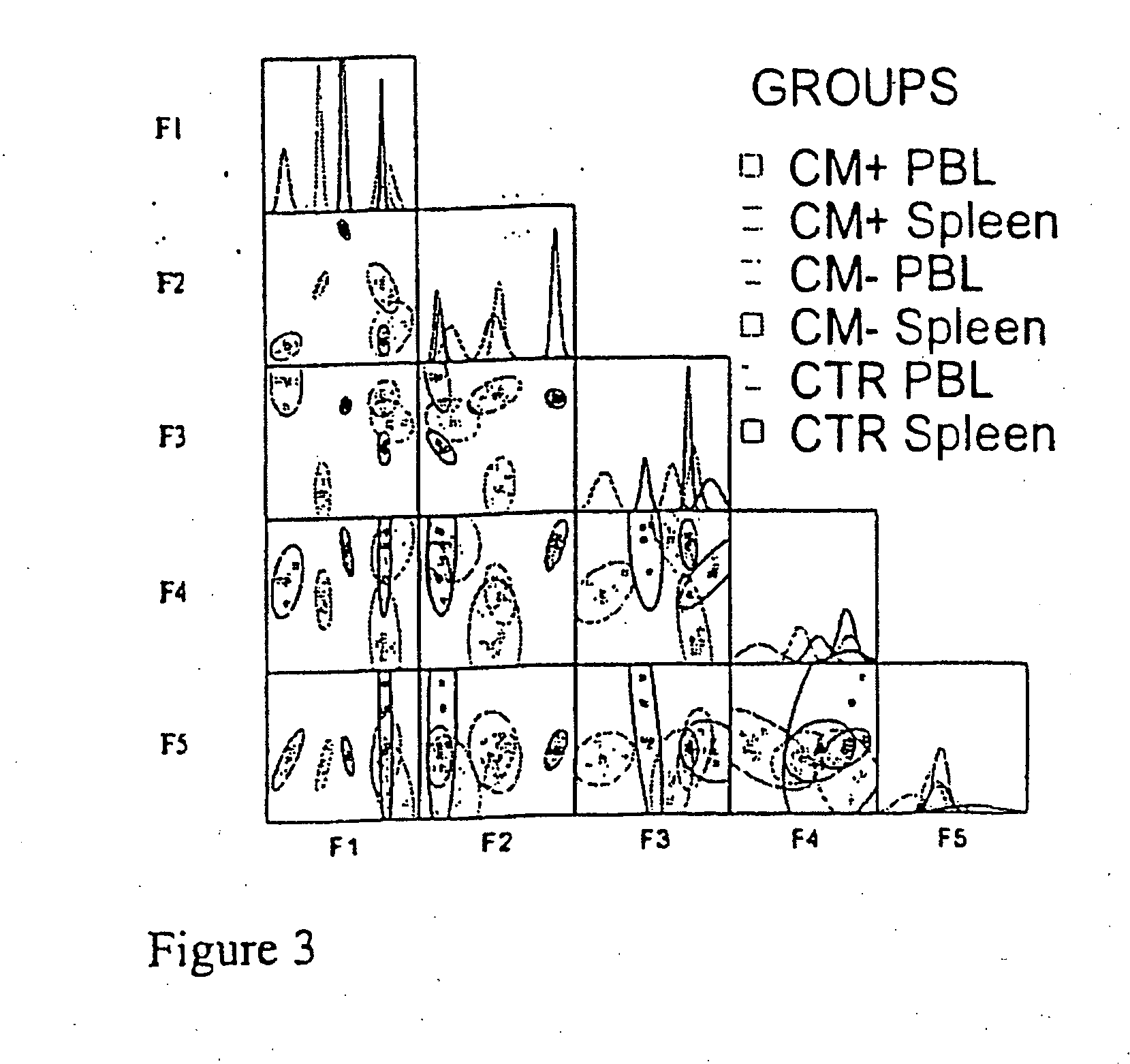
[0135] Eight-week old B10.D2 mice were purchased from Harlan UK Limited. The clone 1.49 L of Plasmodium berghei ANKA (Amani, V., M. I. Boubou, S. Pied, M. Marussig, D. Walliker, D. Mazier, and L. Renia. 1998. Cloned lines of Plasmodium berghei ANKA differ in their abilities to induce Experimental Cerebral Malaria. Infect. Immun 66:4093-4099.) was kindly given by Dr. Walliker (Institute of Genetics, Edinburg, UK) and is maintained in the laboratory on C57BL / 6J female mice. This clone induced in mice a neurological syndrome partly mimicking the one of human CM. Erythrocytic stages of the parasite were cryopreserved in liquid nitrogen as stabilates in Alserver's solution containing 10% glycerol. Infection was induced by intraperitoneal injection of 106 parasitized red blood cells. Between day 7 to day 10 after infection, 90% of the mice developed cerebral malaria characterized by ataxia, paralysis, deviation of the head and convulsions followed by deep coma and death...
example 1.2
Cell Preparation
[0136] Blood was obtained on heparin by retroorbital punction. Mononuclear cells were isolated on ficoll-Hypaque gradient (Pharmacia, France). Spleen was removed and cells suspended in 3% FCS-PBS. Red blood cells were lysed with ammonium chloride buffer (ACK) for five minutes at room temperature. Cell preparations were then washed twice with PBS. Lymphoid cells were counted using Malassez cell in presence of eosin to exclude dead cells.
example 1.3
TCRB Reprtoire
[0137] Total RNA was extracted from more than 90,000 mononuclear cells for each sample using the TRI REAGENT kit (Molecular Research Center, Cincinnati, Ohio). 20 μg of glycogen (Roche, Meylan, France) was used to ensure optimal precipitation of RNA and pellet visualization. Protocols for TCR BV-BC and BV-BJ CDR3 spectratyping have been described previously, which were utilized in the present example (Pannetier, C., M. Cochet, S. Darche, A. Casrouge, M. Zöller, and P. Kourilsky. 1993. The sizes of the CDR3 hypervariable regions of the murine T-cell receptor b chains vary as a function of the recombined germ-line segments. Proc. Natl. Acad. Sci. USA 90:4319-4323.; Pannetier, C., J. Even, and P. Kourilsky. 1995. The Immunoscope technique for analysis of TCR repertoire. In The human antigen T cell receptor: Selected protocols and applications. J. R. Oksenberg, Austin, Tex. 287-325.). BC, BV and BJ primer sequences were those described previously (Pannetier, C., M. Cochet...
PUM
| Property | Measurement | Unit |
|---|---|---|
| DA | aaaaa | aaaaa |
| area | aaaaa | aaaaa |
| TC | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com