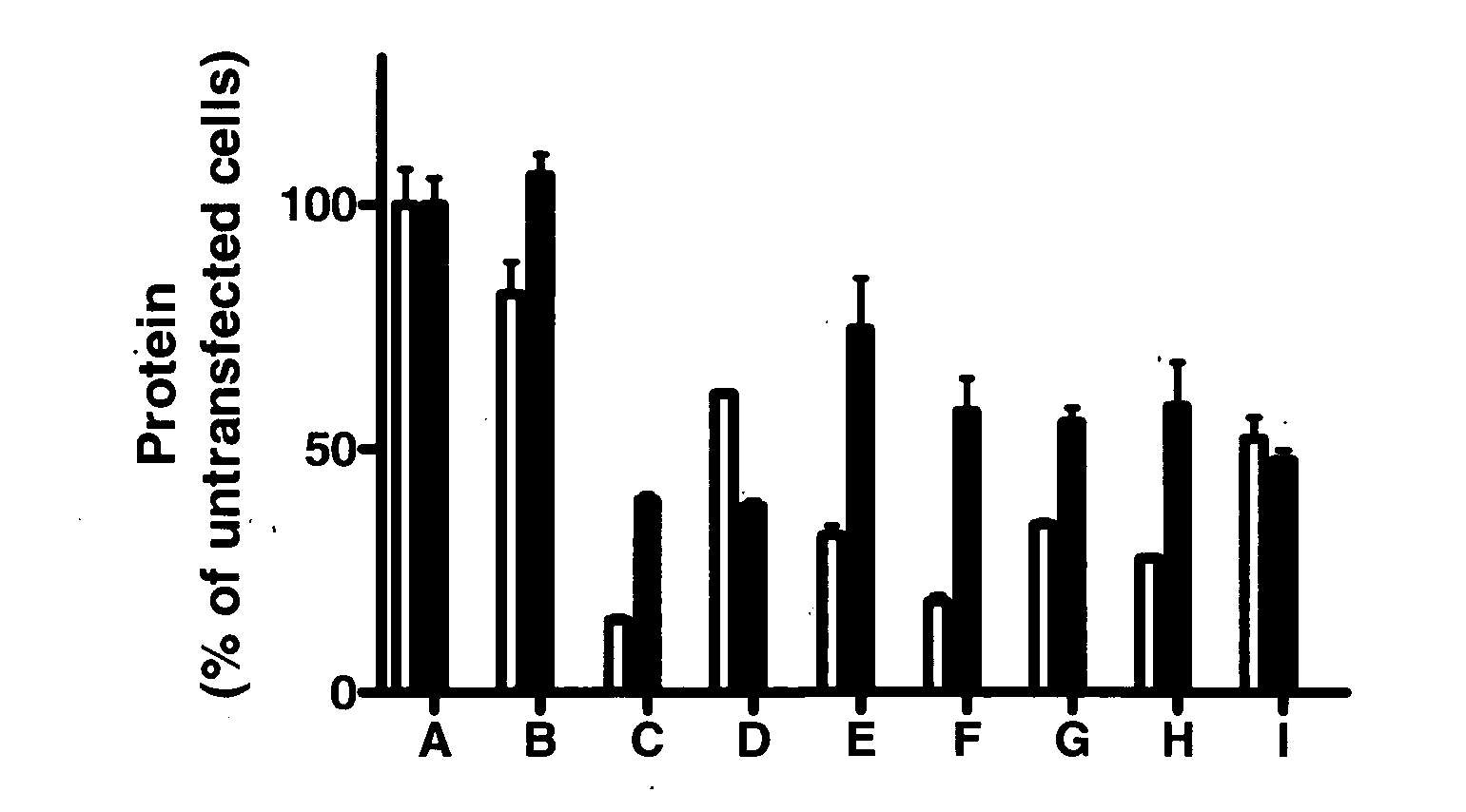Multitargeting interfering RNAs having two active strands and methods for their design and use
a multi-targeting, interfering technology, applied in the direction of viruses/bacteriophages, genetic material ingredients, drug compositions, etc., can solve the problems of reducing the therapeutic effect of single target inhibition, unable to achieve therapeutic efficacy, so as to reduce stress or inflammatory response, increase or decrease the modulation of expression
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
example 1
CODEMIRs to VEGF-A and ICAM-1
[0236] Approaches for the design of multi-targeting of ICAM-1 and VEGF-A with CODEMIRs were considered.
[0237] The entire mRNA sequence for each of VEGF-A and ICAM-1 was used. These sequences were searched to find sequences that are present in the coding strand for one target and the complement of the coding strand for another target. Here, the sequence 5′-AGTGACTGTCAC-3′ (SEQ ID NO: 1) was identified both in the ICAM-1 coding sequence and in the complement of the VEGF-A coding sequence. This sequence was used to design a CODEMIR active against multiple targets, using each strand of the CODEMIR to target at least one of the target RNAs. The sequence identified above and its complement were used as a centrally-located part of a CODEMIR duplex. Each strand of this central duplex was extended in the 5′ direction to provide full complementarity to the corresponding target, whereas each strand was extended in the 3′ direction so as to be complementary to its...
example 2
CODEMIRs to Gluc6P and Inppl1
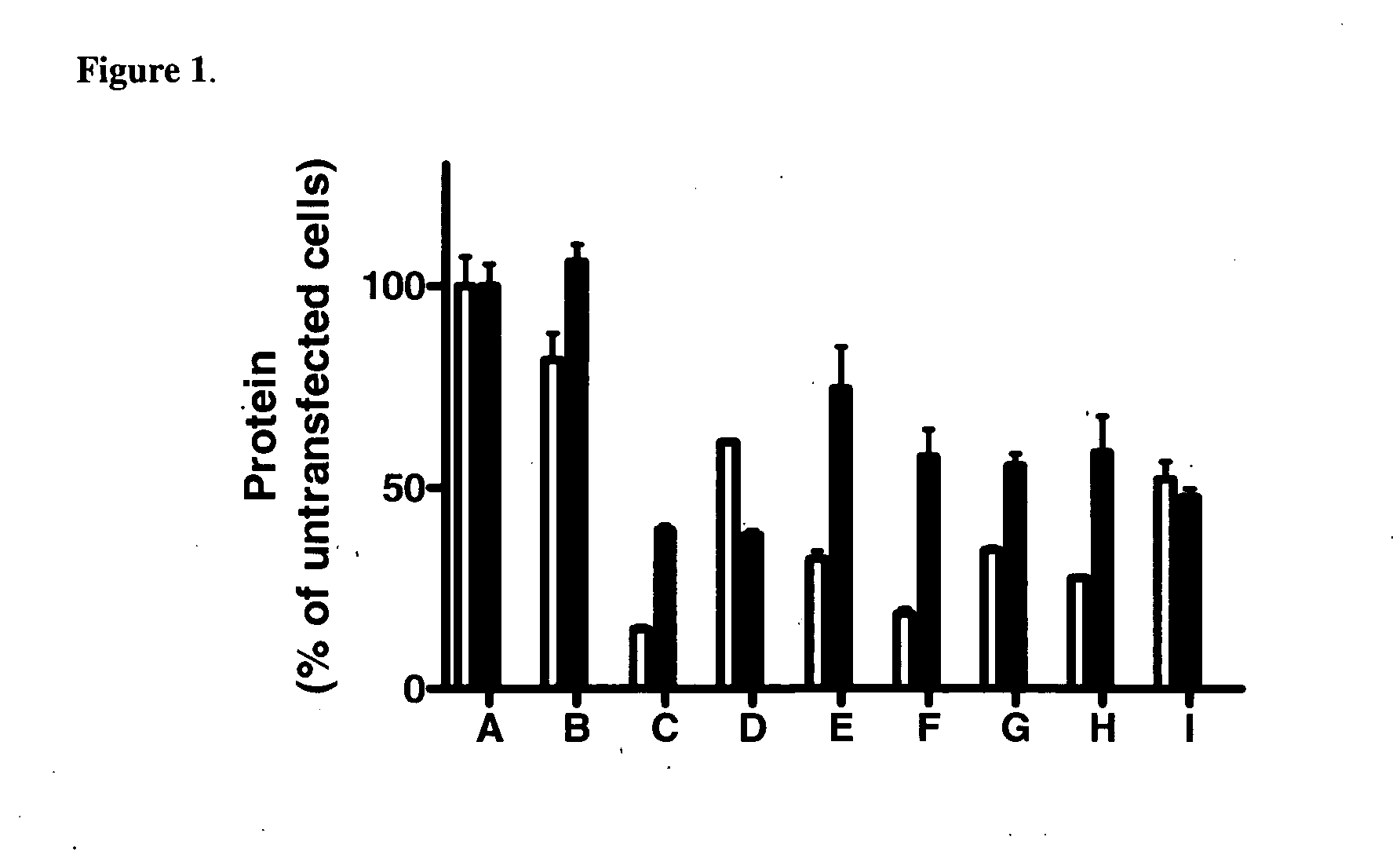
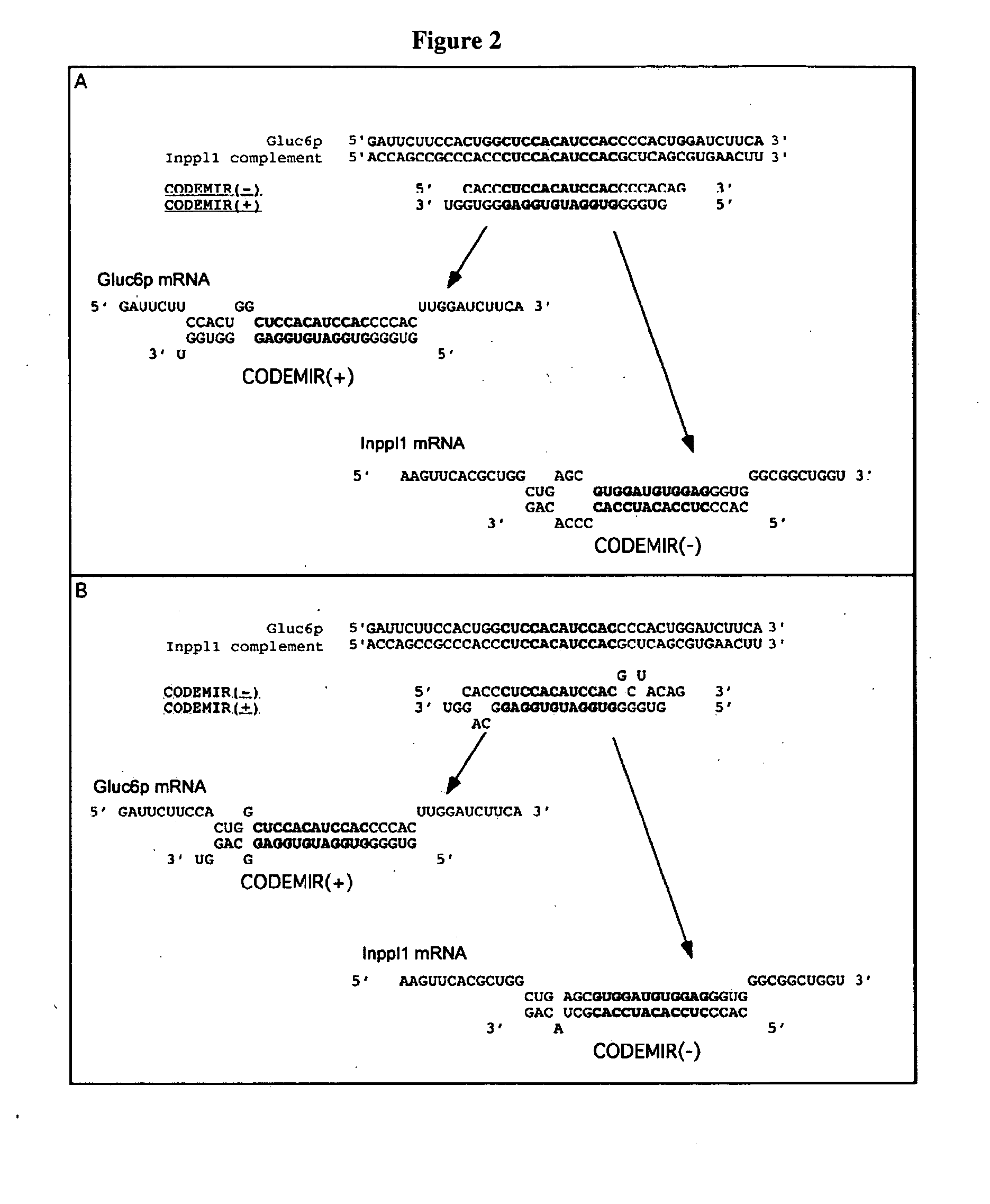
[0249] CODEMIRs may also be suitable for the treatment of complex metabolic diseases such as type 2 diabetes. Two potential gene targets for the treatment of this disease are glucose-6-phosphatase and Inppl1. Full transcript sequences were examined. Candidate CODEMIRs from the best contiguous region of identity are shown for each case in Table 2-1.
[0250] Regions of complementarity between the two targets were found and the two identified seeds (Table 2-1: CUGCCUCGCCCAG (SEQ ID NO: 19) and CUCCACAUCCAC) (SEQ ID NO: 20) were used as the central motifs for two possible CODEMIR duplexes. The latter seed and its complement were extended at their 5′ ends to generate duplexes in which each strand has 5′ complementarity to one of the target sequences (FIG. 2A). This is important because both strands of the CODEMIR duplex are effectors and an increased region of identity of each strand with its target needs to be extended to the 5′ terminus, whereas the less cr...
example 3
VIROMIRs Targeting Multiple Sites Within the HIV Genome
[0252] The invention can be used to target proteins of interest that are likely to be mutated in chronic forms of disease. Mutations may be particularly prevalent in cancer and viral disease in which drug-resistant forms often evolve. In this example, VIROMIRs were designed to target multiple sites in the Human Immunodeficiency Virus (HIV). The requirement for simultaneous mutation at several sites, in order to overcome the effects of such a VIROMIR, is likely to provide a high genetic hurdle to the emergence of resistant viral clones or quasispecies. The genome of the HXB2 strain of HIV I serotype B (GenBank Accession K03455) was used as the principal sequence of interest and was examined with bioinformatics methods detailed elsewhere in this application to find seeds occurring at more than one location. All HIV I clade B isolates in the LANL database as of 1 Aug. 2005 which contain full sequences for any of the GAG, ENV, POL,...
PUM
| Property | Measurement | Unit |
|---|---|---|
| length | aaaaa | aaaaa |
| size | aaaaa | aaaaa |
| step size | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com