Multi-well micropatterning by ablation
a multi-well, ablation technology, applied in the field of multi-well micropatterning by ablation, can solve the problem of limiting the practicality of integration with high throughput formats, and achieve the effect of not increasing experimental complexity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
example 1
Fabrication of the 96-Well Etch Mask
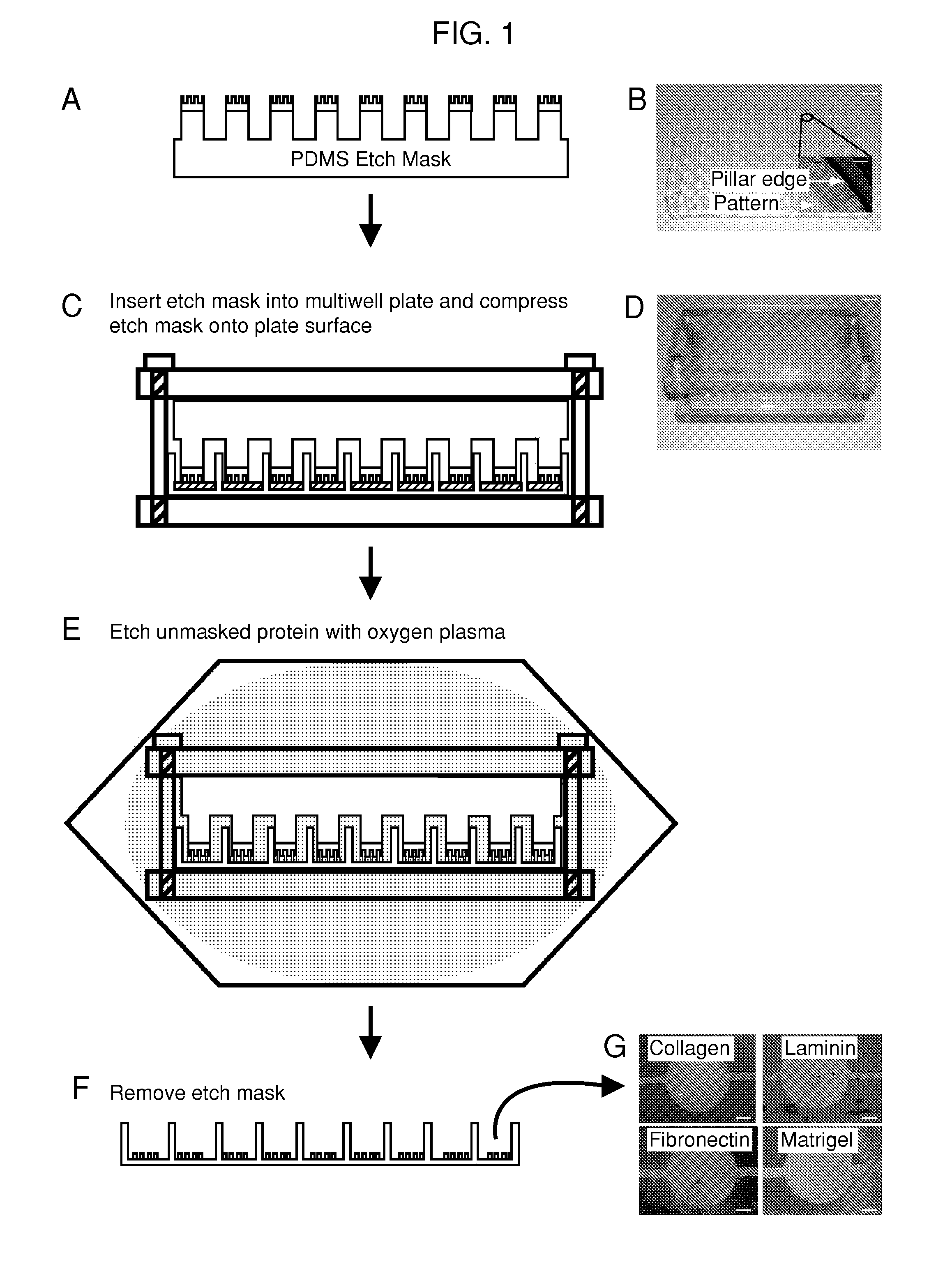
[0128]A mold was machined with the same center to center spacing as a standard 96-well plate and was used to generate the support structure for the 96-well etch mask. This support structure consisted of an array of 96 pillars spaced evenly to allow nesting in a standard 96-well plate. The support structure is molded out of poly(dimethylsiloxane) (PDMS) (sylgard 184, Dow Corning, Midland, Mich.) and prepared using standard techniques. Duffy, D. C.; McDonald, J. C.; Schueller, O. J. A.; Whitesides, G. M. Analytical Chemistry 1998, 70, 4974. In a separate step, mold masters for the micropatterns were fabricated with SU8 photoresist (Microchem, Newton, Mass.) using a high resolution transparency photomask. The molds were fabricated to have 50 μm thick features. These mold masters were coated with a layer of PDMS 2 mm thick The PDMS was peeled from the mold and circles were punched out with a cork borer and glued to the pillars of the PDMS support stru...
example 2
Patterning Protein and Cells in 96-Well Plates
[0129]Biomolecules were physisorbed to each well of a standard multi-well plate (solutions of type-I collagen, fibronectin, Matrigel and laminin at 50 μg / mL in water. Note that 1×PBS may be used instead of water. The multi-well plates were incubated for 1 hour at 37° C. followed by rinsing with water and allowed to air dry. This results in an adsorbed thickness of approximately 150 nm. Gurdak, E.; Dupont-Gillain, C. C.; Booth, J.; Roberts, C. J.; Rouxhet, P. G. Langmuir 2005, 21, 10684-10692. In some cases, micropatterned proteins were fluorescently labeled via incubation (1 hour at room temperature) with Alexa Fluor® 488 carboxylic acid, succinimidyl ester (Invitrogen, Carlsbad, Calif.) dissolved in phosphate buffered saline (PBS) at 20 μg / mL. A single etch mask was inserted into the multi-well plate and compressed in a custom clamp consisting of two blocks of polycarbonate flanking the masked multi-well plate and compression was applie...
example 3
Ablation Quantification
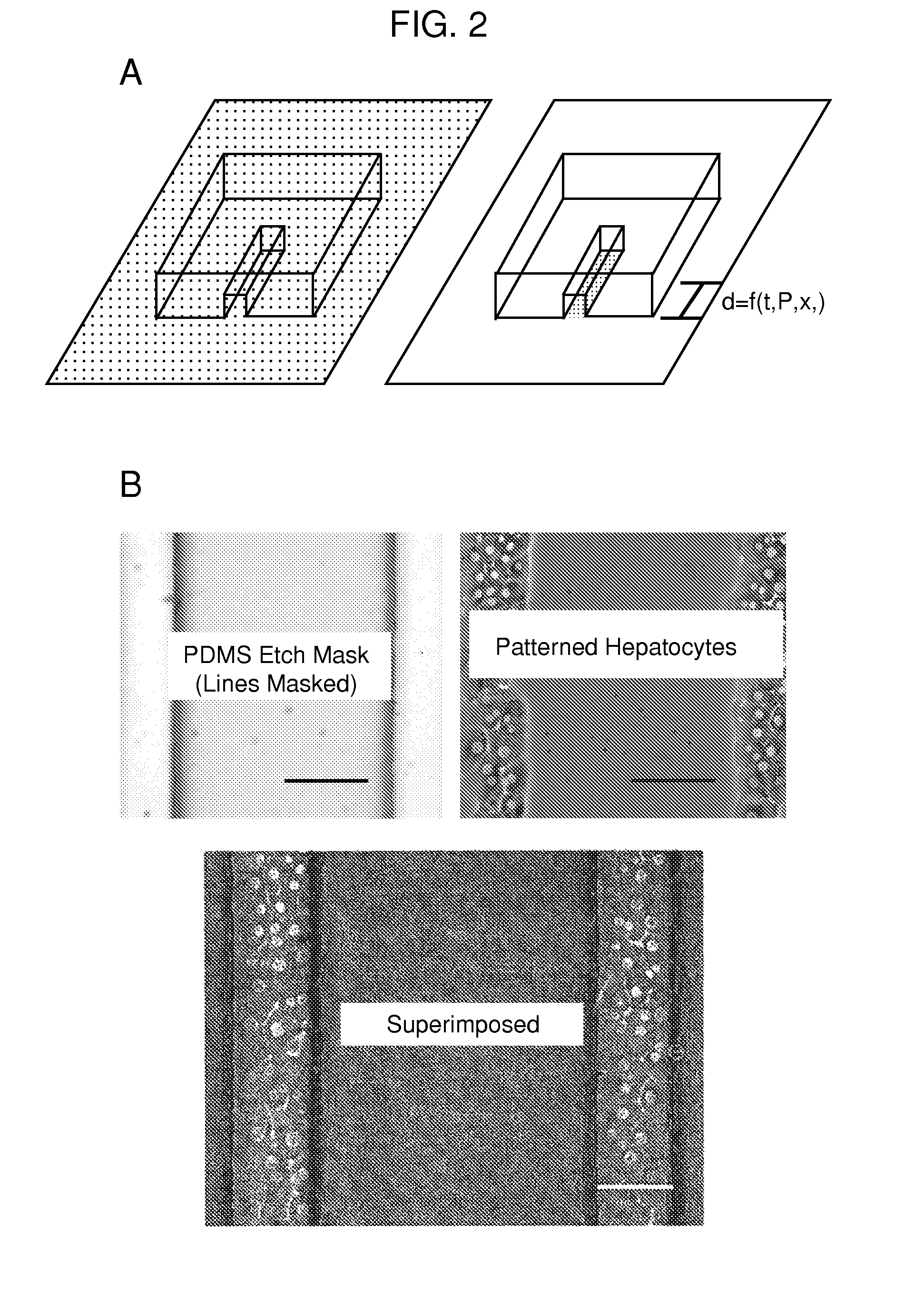
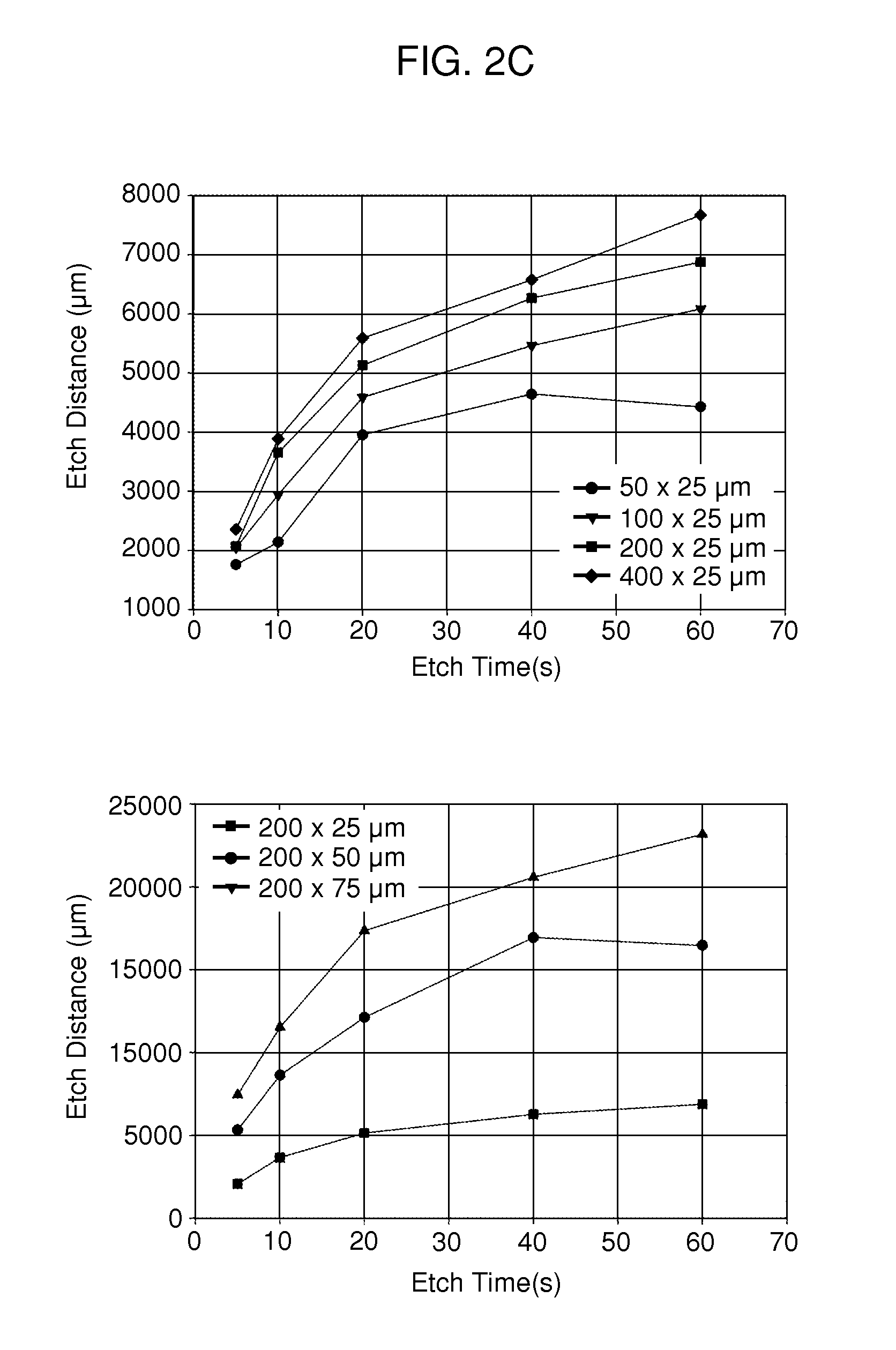
[0131]A PDMS etch mask containing dead-end microchannels with channel widths of 50, 100, 150, and 200 μm repeated for three channel heights of 25, 50, and 75 μm (12 channels total) was used to quantify the etching rate. The mask was placed onto p60 petri dishes physisorbed with type-I collagen and exposed to oxygen plasma for several time points (5, 10, 20, 40, and 60 seconds) with each time point on a separate dish. Primary rat hepatocytes were then seeded onto the patterned dishes and cultured for 24 hours. The hepatocytes specifically attached to regions with type-I collagen, hence plasma ablated distances could be directly correlated to hepatocyte attachment as hepatocytes will not attach to adsorbed collagen substrates from solutions less than 0.5 μ / mL. The distances were measured using Metamorph (Universal Imaging, Sunnyvale, Calif.) and plotted as a function of time.
PUM
| Property | Measurement | Unit |
|---|---|---|
| thick | aaaaa | aaaaa |
| feature size | aaaaa | aaaaa |
| feature size | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com