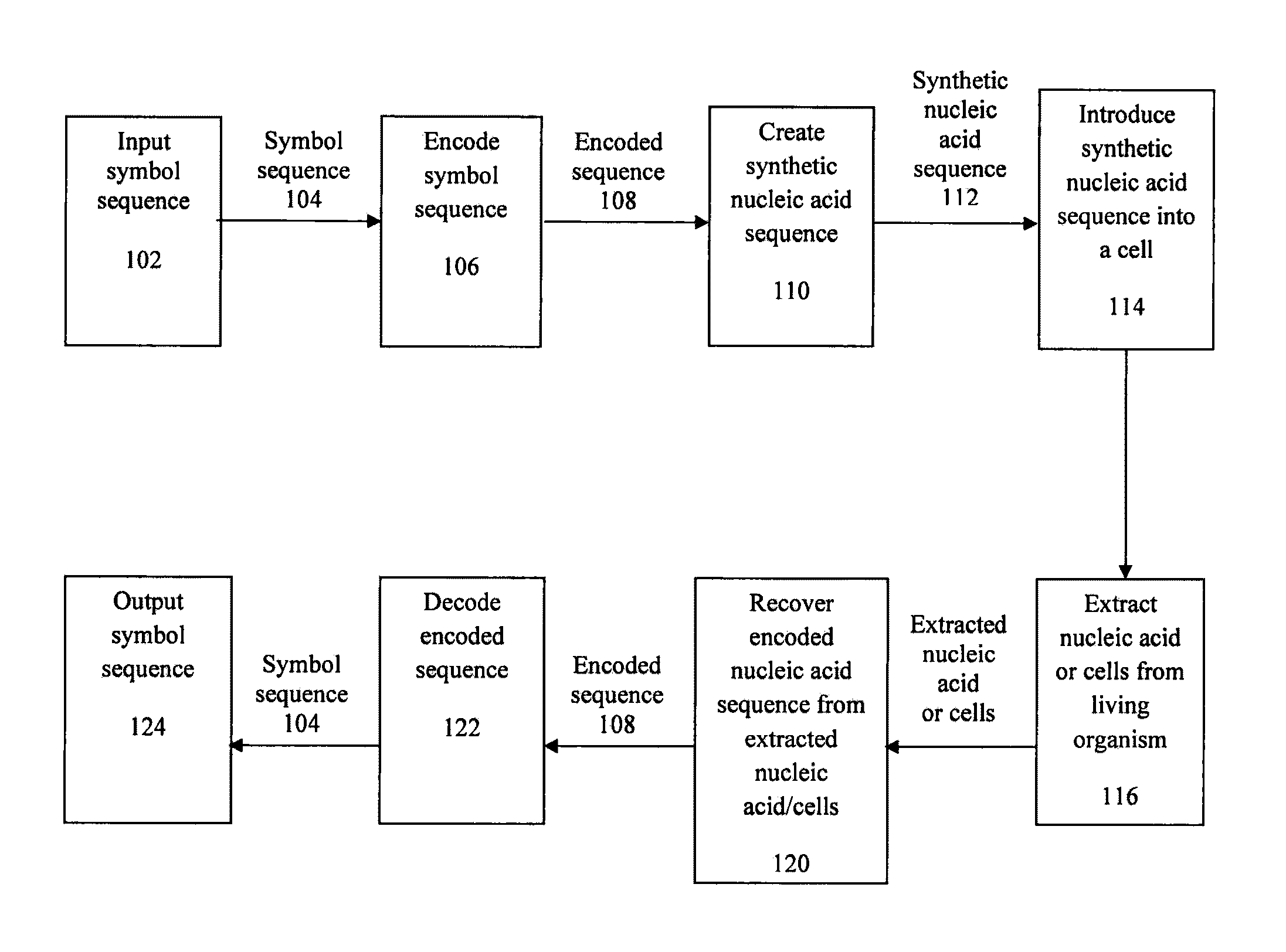
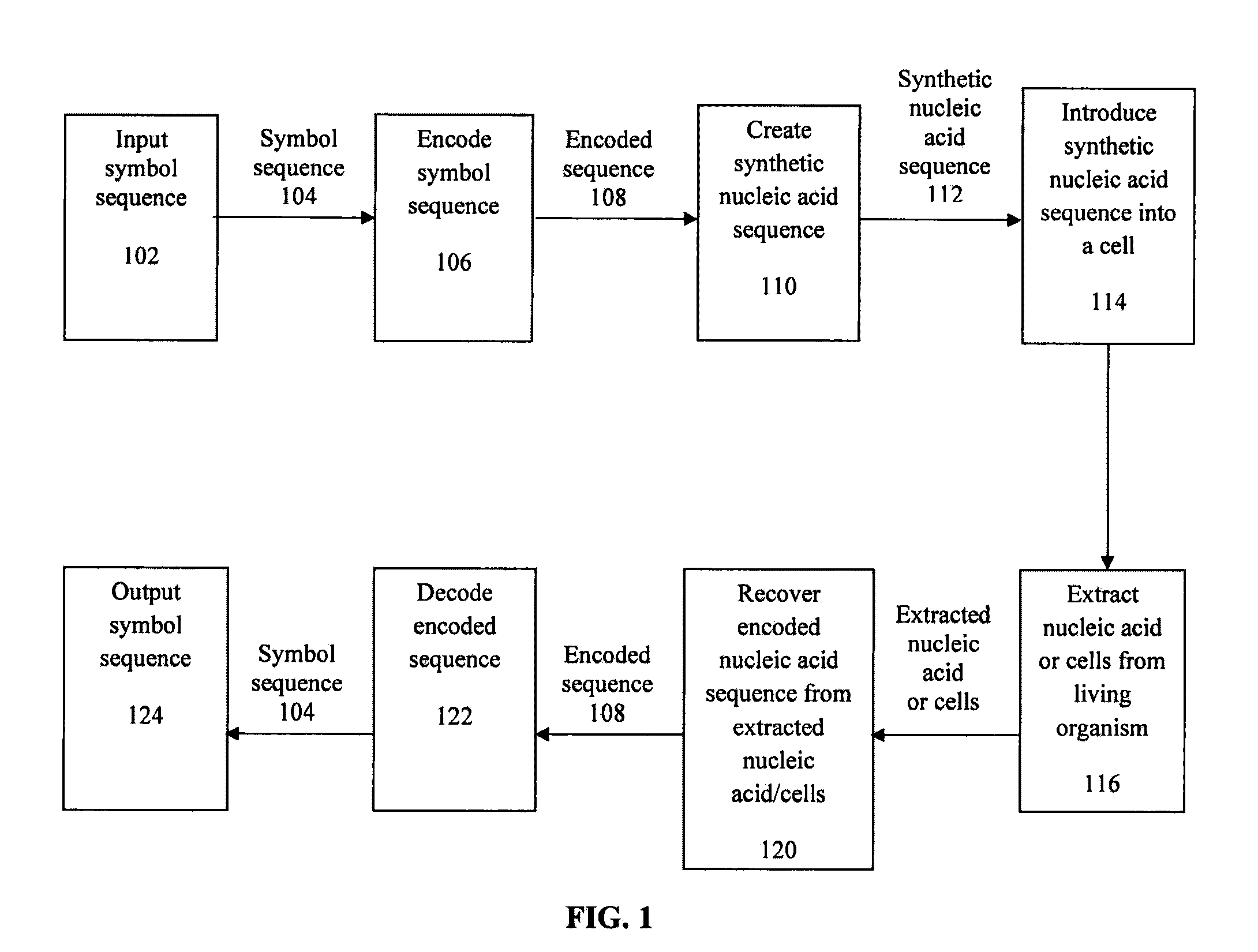
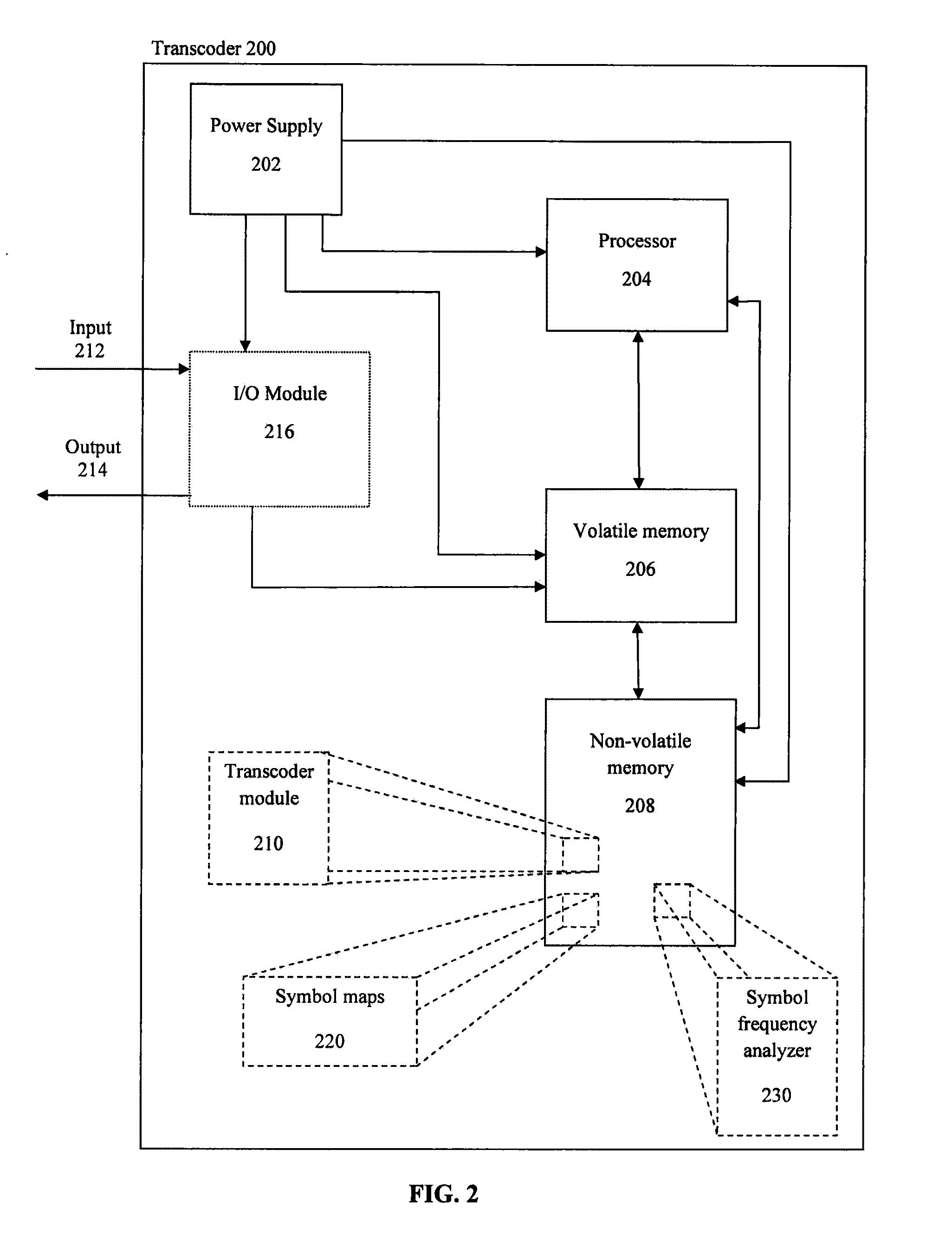
Encoding text into nucleic acid sequences
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
example 1
Encoding Methods
[0234]FIGS. 3 and 8 identify codon identifiers and the respective symbols encoded therefrom. By virtue of the design of the non-genetic message or watermark, the encoded text does not correspond to the sequences of a gene or other biologically active sequence when in the form of a nucleic acid in the cell or organism. The examples provided in the Figures encode all letters in the American English alphabet as well as all 10 numerals and common punctuation marks.
[0235]While the present Figures and Examples are described with respect to the English language, one would comprehend that the coding scheme can be adapted to any reference language as described above.
[0236]An encoded non-genetic message in the nucleic acid sequence is flanked by the sequence 5′-TTAACTAGCTAA-3′ (SEQ ID NO: 1) on both the 5′ and 3′ sides of the watermark since that sequence contains a stop codon in all 6 reading frames.
[0237]To encode a non-genetic message or watermark, one can substitutes in a ...
example 2
[0243]To decode a watermark one performs the same process as encoding as described in Example 1, but in reverse.
[0244]One substitutes in a serial one-to-one manner, each successive three nucleotides of the watermark for their respective human readable symbols of human readable text. These substitutions (performed either by hand or by a computer program) are made such that each human readable symbol is placed to the right of the preceding symbol as one substitutes along the watermark in a 5′ to 3′ direction. This is process of substitution is performed after the sequence 5′-TTAACTAGCTAA-3′ (SEQ ID NO: 1) is removed from both ends of the watermark.
[0245]For example, to decode the sequence 5′-TTAACTAGCTAAGTTTTTTTGCTGCCCGCTTGACTATAGCTGTGCATATCTCTTAC TCGAAATATATAGAACAACATACTACTGTACTCATGAGCTATACTATAAGCTTAA CTATTGTAAATTGTGATAACTTCTTCTGTACGATTAACTAGCTAA-3′ (SEQ ID NO: 9), the first step removes the sequence 5′-TTAACTAGCTAA-3′ (SEQ ID NO: 1) from both ends of the watermark le...
example 3
Synthetic Cells Containing Watermarks
[0251]A 1.08 Mbp Mycoplasma mycoides genome was chemically synthesized, and assembled in yeast as a centromeric plasmid; the genome was isolated as naked DNA and transplanted into Mycoplasma capricolum to create a new bacterial cell controlled only by the synthetic genome.
[0252]Described in International Patent Application PCT / US10 / 35490 is the design, synthesis and assembly of the 1,077,947-bp Mycoplasma mycoides JCVI-syn1 genome from 1,078 1-kb synthetic DNA cassettes. The assembly was facilitated by in vitro and in vivo assembly methods. Cassettes in sets of ten were assembled by yeast recombination and propagated in a yeast / Escherichia coli shuttle vector. The 10-kb assemblies were recombined in sets of ten to produce 100-kb assemblies. The resulting eleven 100-kb assemblies were recombined in a single final step into the complete genome. A yeast clone bearing the synthetic genome was selected and confirmed by multiplex PCR and restriction an...
PUM
| Property | Measurement | Unit |
|---|---|---|
| Biological properties | aaaaa | aaaaa |
| Nucleic acid sequence | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com