Masp isoforms as inhibitors of complement activation
a technology of complement activation and isoforms, which is applied in the field of new ficolin-associated polypeptides, can solve problems such as increasing the risk of infections
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
example 1
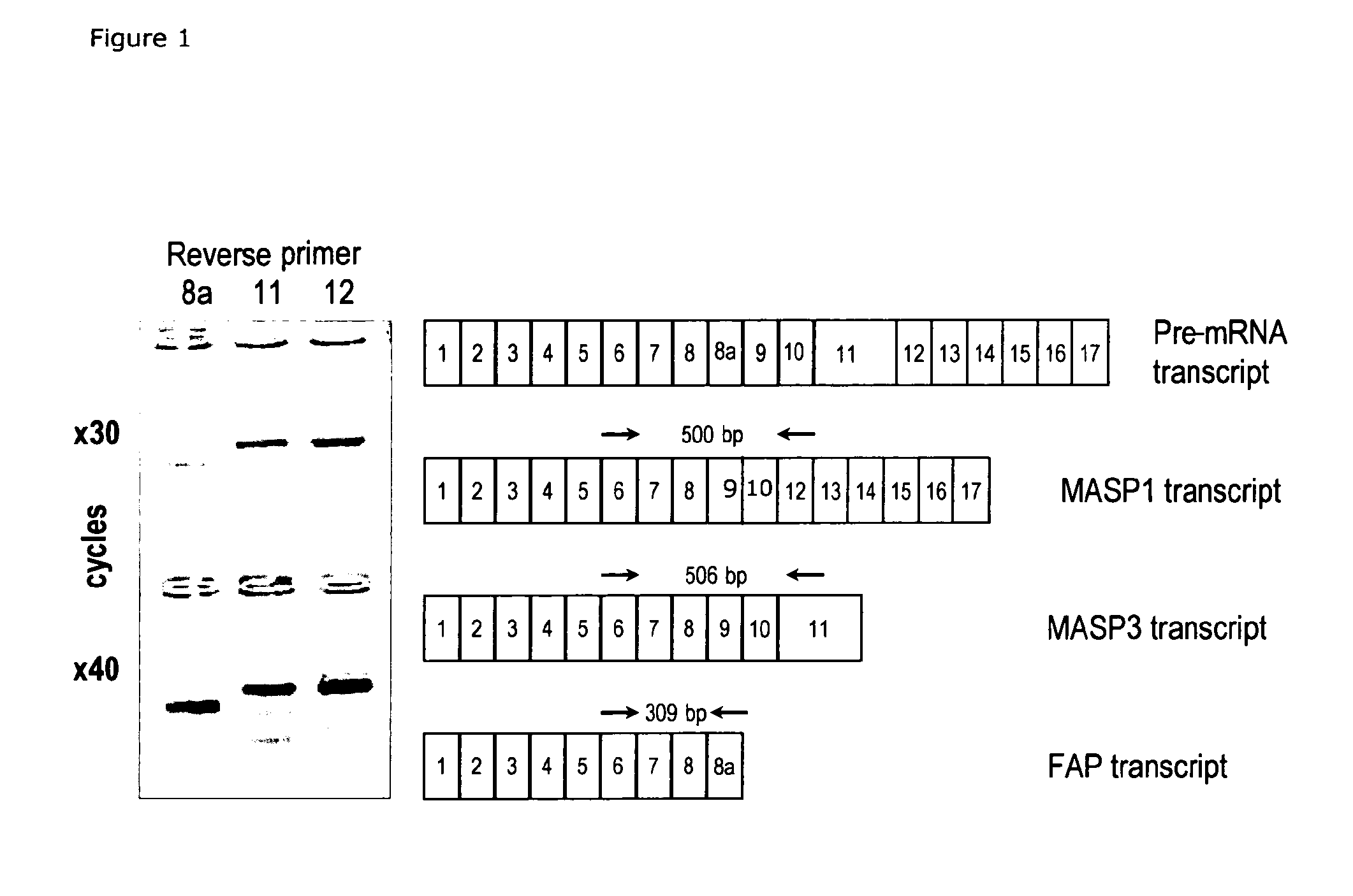
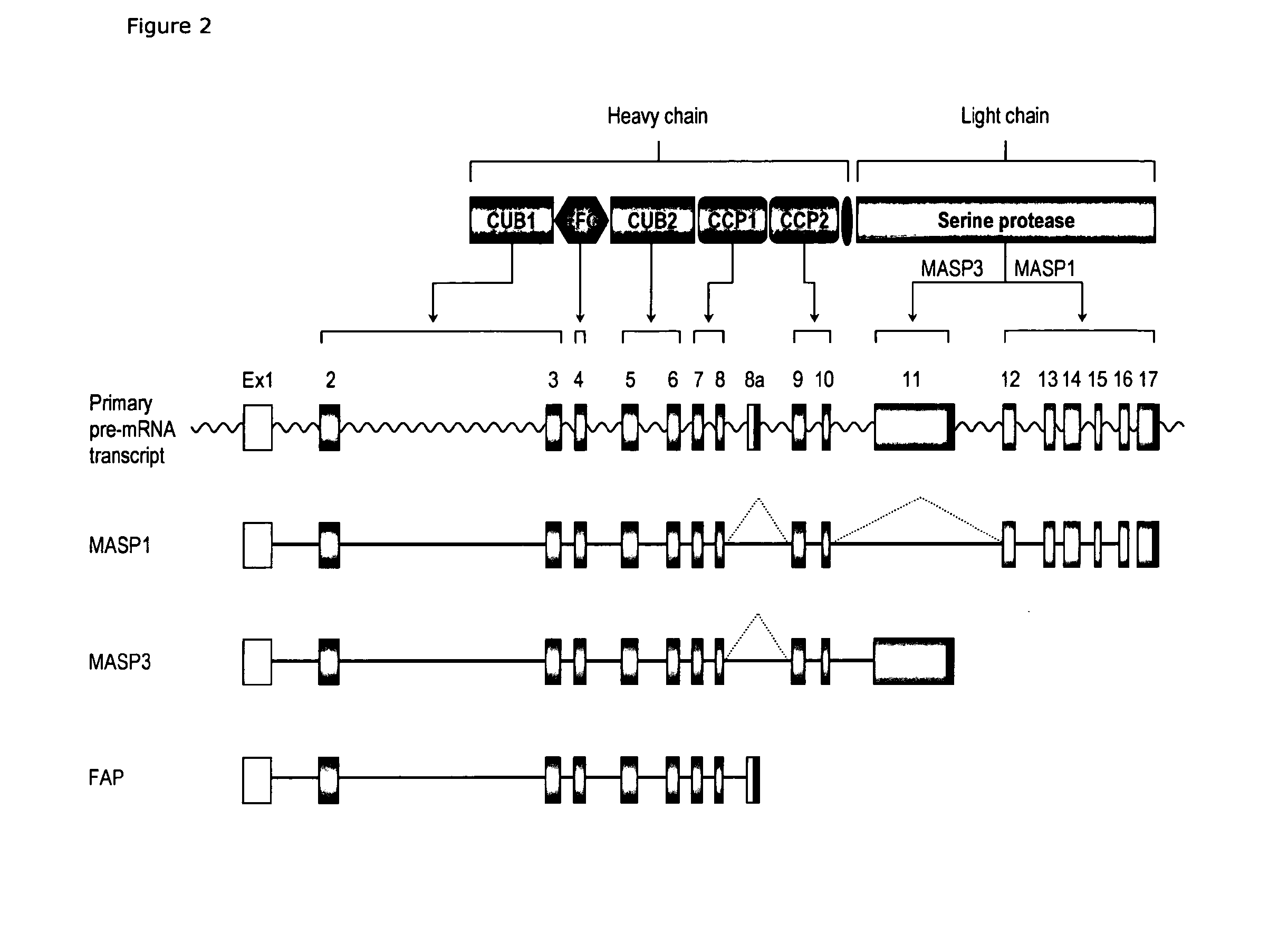
[0316]Detection of Alternative Transcription of the MASP1 Gene
[0317]Methods: In order to detect the three transcript variants of MASP1: MASP1, MASP3 and FAP, specific primers for each variant were design. PCR was set up with a common forward primer in exon 6 (5′-gcacccagagccacagtg-3′) and specific reverse primers: MASP1 in exon 12 (5′-gccttccagtgtgtgggc-3′), MASP3 in exon 11 (5-gccttccagagtgtggtca-3′) and FAP in exon 8a (5′-cgatctggagagcgaactc-3′) (FIG. 1). PCR amplifications were carried out in 20-μl volumes containing: 50 ng liver cDNA (Clontech), 0.25 μM of each primer, 2.5 mM MgCl2, 0.2 mM dNTP, 50 mM KCl, 10 mM Tris.HCl, pH 8.4, and 0.4 units of Platinum Taq DNA polymerase (Invitrogen). The PCR reactions were performed at the following cycling parameters: 10 min 94° C., 30 or 40 cycles (30 sec 94° C., 50 sec 58° C., 90 sec 72° C.), 10 min 72° C. Samples were analysed on 2% agarose gels.
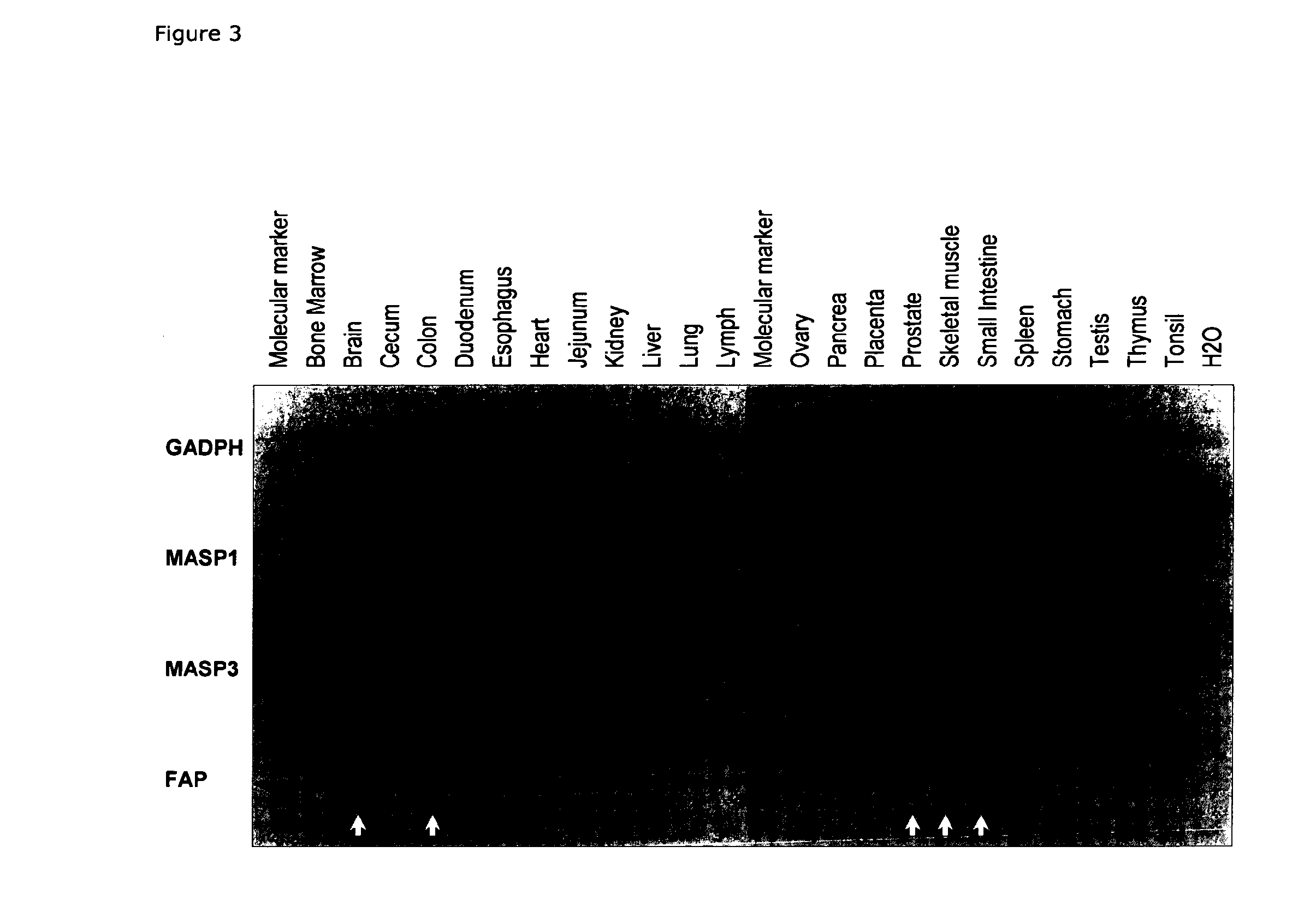
[0318]Results: Alternative transcription of the MASP1 gene was detected in liver cDNA. The MA...
example 2
[0325]Immunoprecipitation.
[0326]Specific immunoprecipitation of MAP-1 from serum was performed with the MAP-1 specific mAb 20C4 (raised against the 17 MAP-1 specific C-terminal peptide) or mAb 8B3, a monoclonal antibody reacting against the common heavy chain of MASP-1 / 3 used as control precipitation antibody. A total of 10 μg of anti MAP-1 or MASP-1 / 3 antibody was allowed to bind to sheep anti mouse or rabbit IgG Dynabeads (M-280, cat. 112.02D / 112.04D, Dynal / Invitrogen). After a washing step the beads were applied to a pool of normal human serum (diluted 1:1 in TBS) and incubated end over end for 1 hour at 4° C. After final washing steps and magnetic separation the beads were boiled in SDS loading buffer and subjected to SDS-PAGE and western blotting probed with antibodies to MAP-1, MBL, and Ficolin-3.
[0327]The same precipitation procedure as described above was performed with mAbs to MBL (Hyb 131-11, Bioporto, Denmark), Ficolin-2 (FCN219) and Ficolin-3 (FCN334). To compensate for ...
example 3
[0341]Determining serum concentration and association properties of the novel MBL / Ficolin associated protein 1 (MAP-1).
[0342]A full-length non-tagged recombinant constructs of MAP-1 was generated and stably expressed in CHO-DG44 cells. Specific monoclonal antibodies against MAP-1 were raised. Also a quantitative ELISA for MAP-1 serum measurements was established and the associations between serum MAP-1 and Ficolin-2, -3 and MBL was examined by ELISA and density gradient fractionation.
[0343]Recombinant Proteins
[0344]Full length constructs of non-tagged human MAP-1 was expressed in CHO-DG44 cells as described elsewhere (Hummelshoj et al., Mol Immunol 44, 401-11, 2007; Larsen et al., J Biol Chem 279, 21302-11, 2004; Ma et al., 2009 J Biol Chem, October 9; 284(41)) with the modifications that PowerCHO1 serum-free medium (Lonza, Vallensbaek / Denmark, www.lonza.com) was used as the expression medium. We used antibody affinity purification to purify rMAP-1 as described previously (Skjoedt e...
PUM
| Property | Measurement | Unit |
|---|---|---|
| Molar density | aaaaa | aaaaa |
| Molar density | aaaaa | aaaaa |
| Composition | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com