Immunogenetic restriction on elicitation of antibodies
a technology of immunogenetic restriction and antibody elicitation, which is applied in the field of influenza neutralizing antibodies, can solve the problems of low detection concentration of sbnabs and no approach that can evaluate the ability of influenza vaccines, and achieve the effect of improving the neutralization capacity or affinity of antibodies
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
example 1
ethods
[0248]Described herein are the general methods and assays used in the working examples.
[0249]Materials
[0250]The anti-HA antibodies F10, A66, G17, and D8 were previously described in the study of Sui et al (1) and International Application No. WO 2009 / 079259, herein incorporated by reference in their entireties. The mAbs CR6261 and CR6331 were synthesized by Genewiz, North Brunswick, N.J. Recombinant HA of H5VN04 was produced as described (1). A / California / 04 / 2009 (H1CA0409) and A / Singapore / 1 / 57 (H2 SIN 57) recombinant HAs were supplied by Biodefense and Emerging Infections Research Resources Repository (BEI Resources)
[0251]Cloning of Antibody Variants
[0252]IGHV1-69*01 germline V-segment was synthesized by Geneart (Regensburg, Germany). The germline variant of IGHV1-69 / F10 was constructed by ligating the IGHV1-69*01 gene (NcoI 5′, BssHII 3′) with F10 gene segment that included the CDR-H3+ light chain (BssHII 5′ NotI 3′) into the pET22b vector, which was digested with NcoI-NotI....
example 2
ation of Anchor Residues in the Heavy Chains of HVI-69-SBNABS
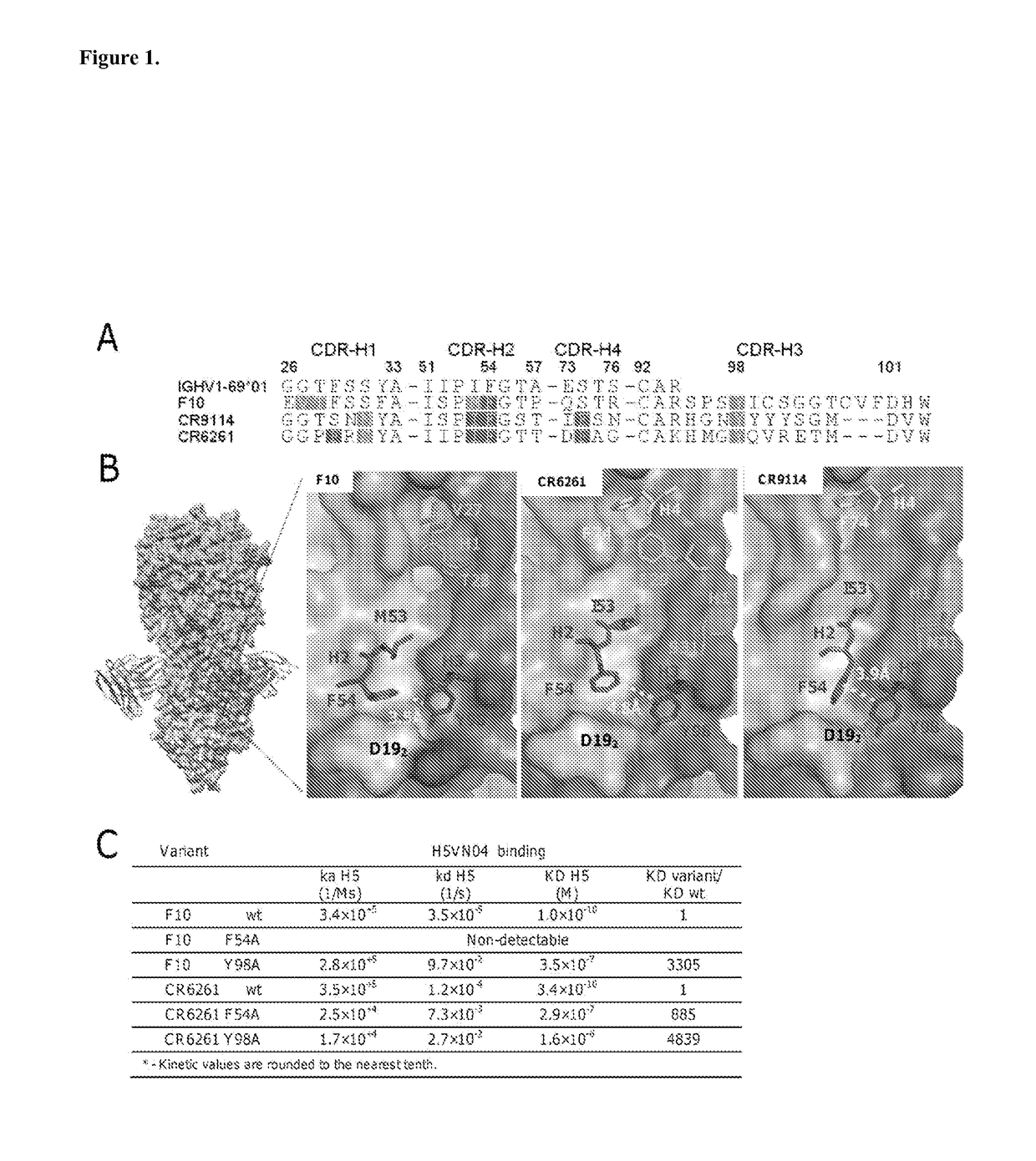
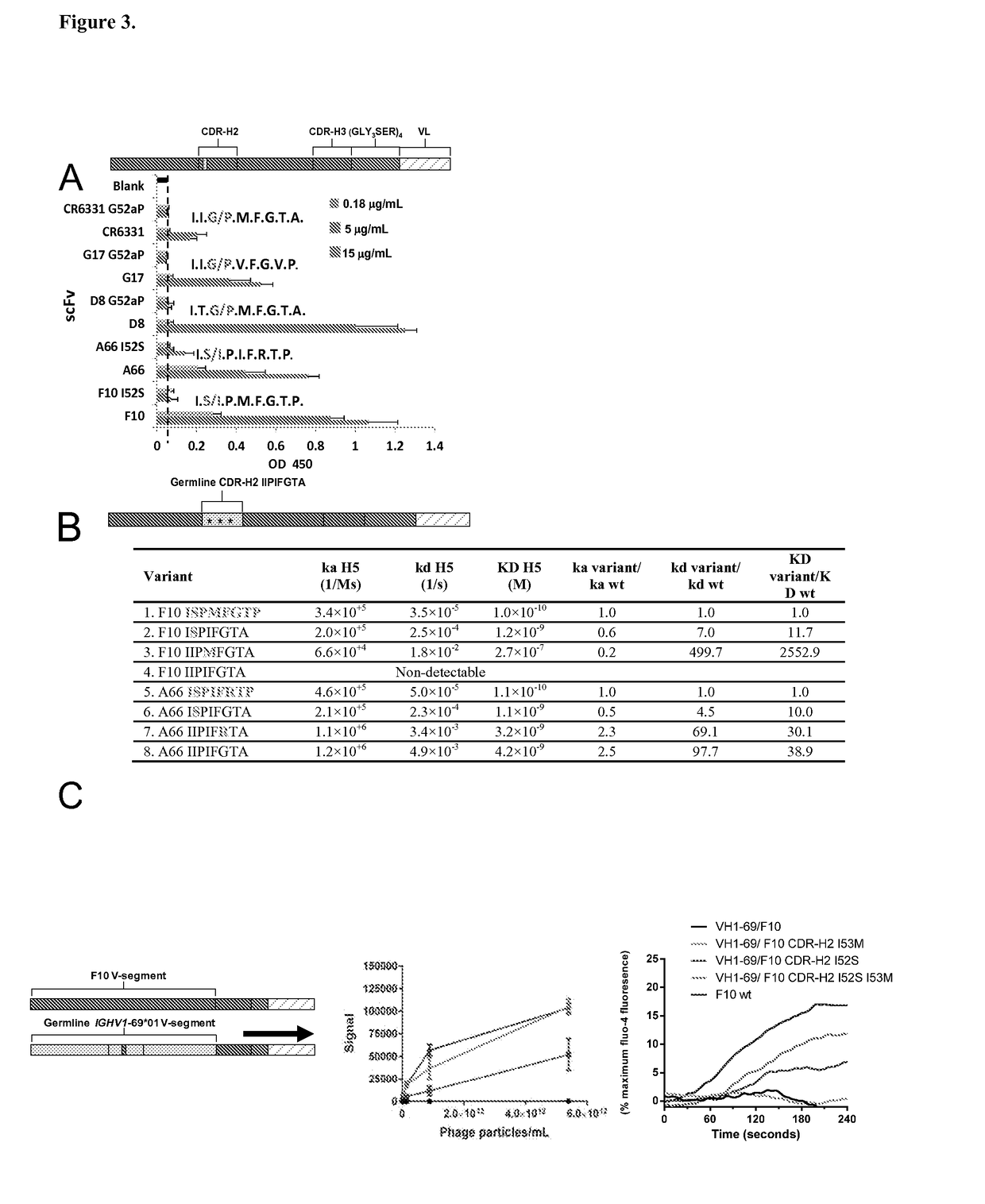
[0284]The co-crystal structures of the HV1-69-sBnAbs, F10 (1), CR6261 (3), and CR9114 (5) with H5VN04 established that binding is mediated exclusively by the IGHV1-69 heavy chains. Estimates of the binding free energy contributions for heavy chain CDR residues using ANCHOR server (17) (FIG. 1A) have identified three common anchor points: a hydrophobic residue at CDR-H2 position 53 (generally Ile / Met), a Phe at CDR-H2 position 54, and a Tyr residue at CDR-H3 position 98. Structural analysis shows (FIGS. 1B and 6) that the common aromatic pair of CDR-H2 Phe54 and CDR-H3 Tyr98 pack closely together (˜4 Å) in order to bind to adjacent pockets formed by elements of the fusion peptide. Tyr98 makes both hydrophobic interactions as well as a strong H-bond with the fusion peptide (the main chain carbonyl of Asp192), and adopts a single conformation in the 3 known structures. The side-chains of Phe54 converge in one location, packin...
example 3
nalysis of Somatic Mutations in Rearranged IGHV1-69 Genes of the SBNABS
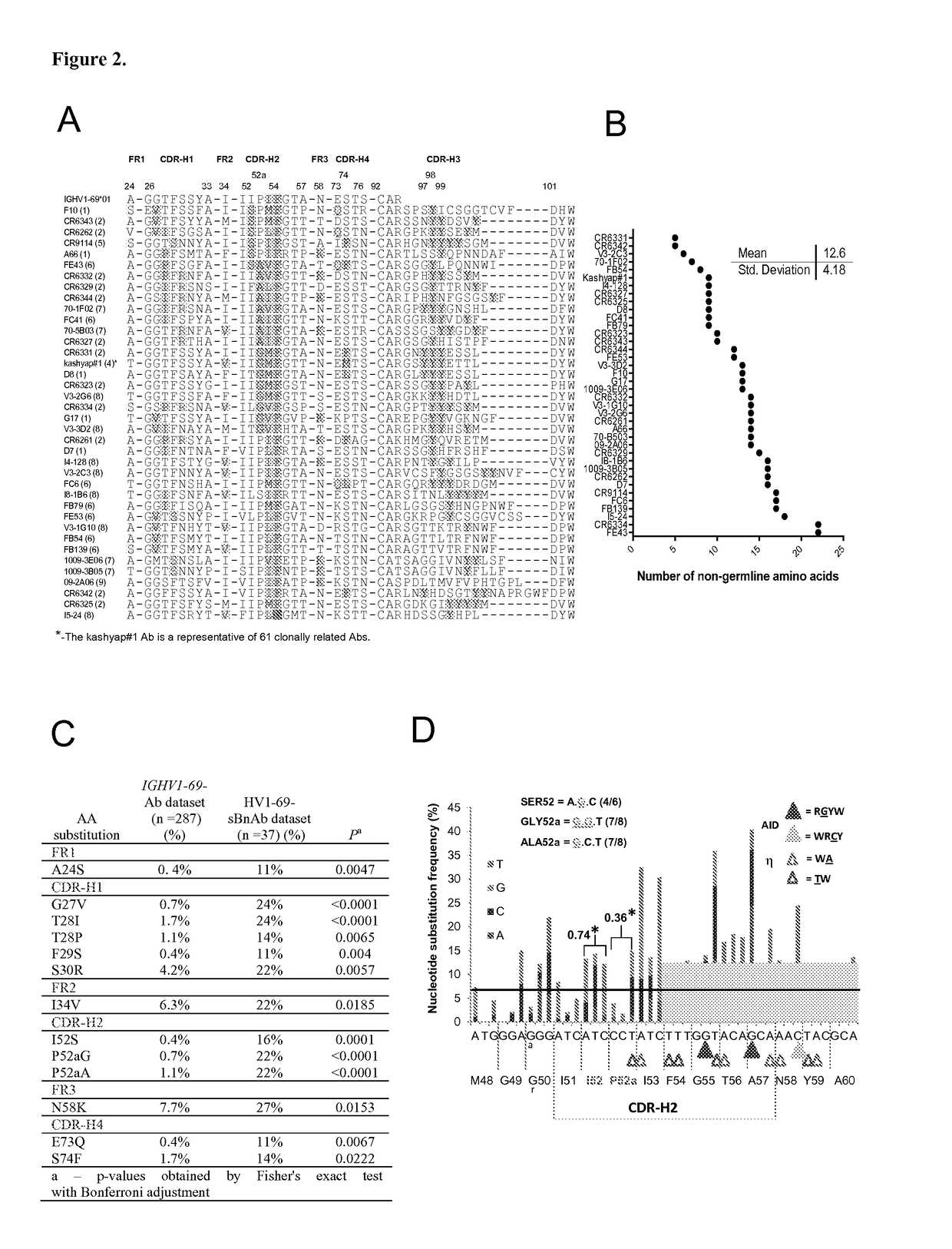
[0286]A mean of 12.6±4.2 V-segment substitutions are found among the published HV1-69-sBnAbs, ranging from 5 in CR6331 / CR6432 to 22 in FE43 / CR6334 (FIG. 2B), which is similar to the average range for rearranged IGHV genes (21). This distribution indicates the pathways for potent HV1-69-sBnAbs formation do not necessarily require multiple maturation events, but rather the incorporation of key residues. Further examination of the V-segments of HV1-69-sBnAbs revealed common substitutions such as the hydrophobic residue at position 74 (in CDR-H4) and changes in CDR-H1 and CDR-H2 (FIG. 2A). To investigate which of these substitutions are unique for HV1-69-sBnAbs, we compared the frequency of specific somatic mutations in the HV1-69-sBnAbs V-segments with the identical mutations in a control set of unique Ab sequences derived from the IGHV1-69-51p1 germline group (IgBlast, n=287). Thirteen HV1-69-sBnAb distinctive V-se...
PUM
| Property | Measurement | Unit |
|---|---|---|
| Digital information | aaaaa | aaaaa |
| Composition | aaaaa | aaaaa |
| Therapeutic | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com