Method of identifying invasion of south American glim ant and its nucleic acid sequence, probe and reagent kit
A nucleic acid sequence, red fire ant technology, applied in the field of molecular biology, can solve the problems of time-consuming and difficult to fully guarantee the accuracy, and achieve the effect of eliminating experimental operation errors, small sample volume, and strict inspection standards.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0039] Example 1 Preparation of invasive South American red fire ants rDNA spacer (ITS region) nucleic acid sequence
[0040] Take a specimen of RIFA preserved by soaking in alcohol and put it into a 1.5ml centrifuge tube. Cooled with a small amount of liquid nitrogen, frozen and brittle. Grind slightly to crush the sample. Add 500ul extraction buffer (20mM Tris-Cl pH 8.0, 20mM EDTA pH 8.0, 400mM NaCl, 1% SDS), proteinase K (20mg / ml) 10ul. Incubate at 55°C and digest with shaking (750rpm) for more than 5 hours. Extraction with phenol:chloroform:isoamyl alcohol (25:24:1). Ethanol precipitation overnight. Redissolve with appropriate amount of TE to obtain the total DNA of RIFA invading South America.
[0041] Then carry out the PCR of ITS region, the primers used are respectively LH2 (5'CGTAGGTGAACCTGCGGAAGGATC 3') and Sm73 (5'TTCGCTCGCCGTTACTAGGGGAATC 3'). Take a 200ul PCR tube, add 20ul PCR reaction solution (which contains 2.0mM MgCl 2 , 200uM dNTP, 10×Taq DNA buffer 2...
Embodiment 2
[0042] Design and preparation of embodiment 2 specificity probe
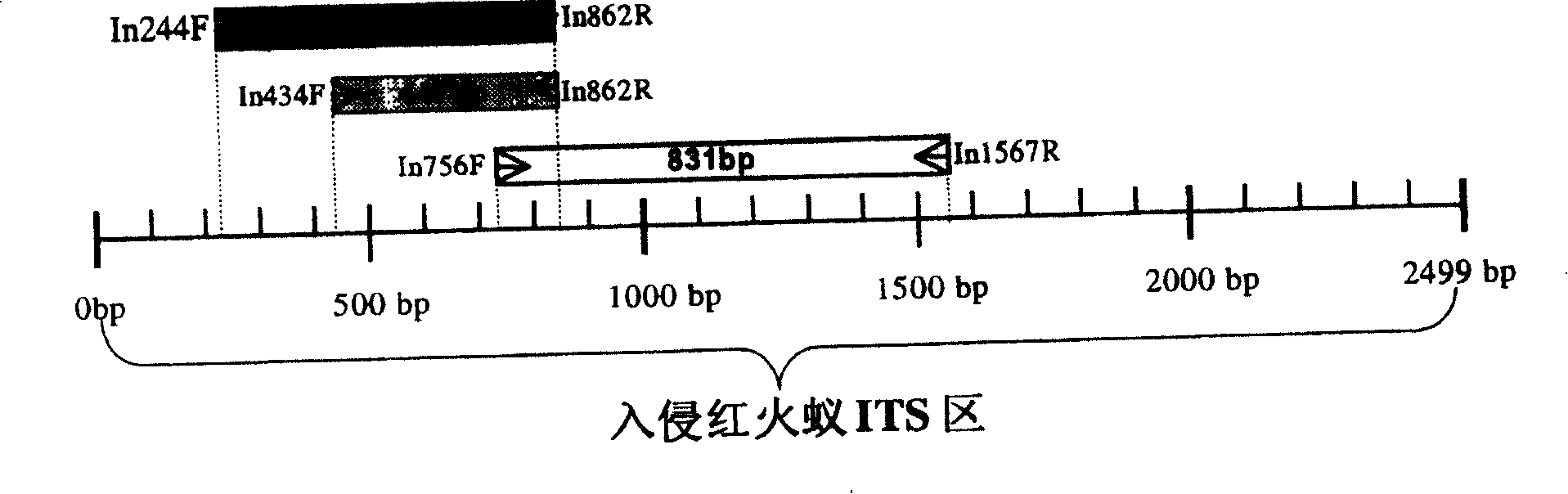
[0043] On the basis of obtaining the nucleic acid sequence of the rDNA spacer of the invasive South American red imported fire ants, it was aligned and compared with the homologous sequences of other ant species, and a specific Probes (oligonucleotide fragments) with good properties. According to the sequence of the oligonucleotide fragments, chemical synthesis was carried out on a commercial DNA synthesizer. Five types of nucleic acid molecular probes were synthesized, as follows:
[0044] In244F: 5'ATCTTGGTGGAATTGACGAC 3'
[0045] In434F: 5'GACGGGAGGGAAGAAAAGAC 3'
[0046] In756F: 5'TGCTTAGCACGCTGGGAT 3'
[0047] In862R: 5'AGAGACCGCCGAGAAGTGC 3'
[0048] In1567R: 5' ACAACCGCGAGAGGGACTAT 3'.
[0049] The schematic diagram of the mutual position and direction relationship of the specific probe and PCR-specific primer combination in the ITS region DNA sequence of the invasive South American red imported fir...
Embodiment 3
[0051] The identification (conventional PCR method) of embodiment 3 invading South American red fire ants
[0052] 1. Extraction of total DNA from the sample.
[0053] Take one of each ant sample to be tested and put it into a 1.5ml centrifuge tube respectively. Add a small amount of liquid nitrogen to cool, freeze and crisp. Grind slightly to crush the sample. Add 500ul extraction buffer (20mM Tris-Cl pH 8.0, 20mM EDTA pH 8.0, 400mM NaCl, 1% SDS), proteinase K (20mg / ml) 10ul. Incubate at 55°C and digest with shaking (750rpm) for more than 5 hours. Extraction with phenol:chloroform:isoamyl alcohol (25:24:1). Ethanol precipitation overnight. Redissolve with an appropriate amount of TE to obtain the total DNA of the sample to be tested.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com