Inhibitors of MEMAPSIN2 and use thereof
A technology of MMI-026 and MMI-025, which is applied in the field of treatment and/or prevention of Alzheimer's disease and treatment of Alzheimer's disease patients, and can solve problems such as inability to prepare active enzymes
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0133] Example 1 Proteolytic activity and the fragmentation site priority of recombinant memapsin2
[0134] To confirm the specificity of memapsin2, the amino acid sequence around the APP proteolytic cleavage site was determined. Recombinant pro-memapsin2-T1 was incubated in pH 4.0, 0.1M sodium acetate solution for 16 hours at room temperature to generate autocatalytic cleavage. Products were analyzed using SDS-polyacrylamide gel electrophoresis. Many bands with molecular weights smaller than pro-memapsin2 were found. The bands were blotted onto PVDF membranes. Four swimming bands were selected, and their N-terminal sequences were determined in a protein sequencer. The N-terminal sequence of these bands identifies the location of the pro-memapsin2 proteolytic cleavage site.
[0135] In addition, the oxidized beta chain of bovine insulin and two different synthetic peptides were used as substrates for memapsin2 to determine the extent of other hydrolysis sites. Put prome...
Embodiment 2
[0141] Example 2 Activation and enzyme kinetics of pro-memapsin2
[0142] Pro-M2 at 22°C pd Incubate in 0.1M sodium acetate solution at pH 4.0 for 16 hours to transform into M2 by autocatalysis pd . To determine the initial rate of hydrolysis, two synthetic peptides were mixed with pro-M2 pd Cultured in 0.1M sodium acetate solution at pH 4.0, and maintained for 2-18 hours. Cultured samples were analyzed using liquid chromatography (LC) and mass spectrometry (MC) to identify hydrolysates. For kinetic studies, identified HPLC (Beckman System Gold) product peaks were integrated for quantitative analysis. Determination of K of Sensenin 1 and Swedish APP Peptides (Table 1) Using Static Kinetics m and K cat value. Since the K of the APP peptide cannot be accurately determined by standard methods m and K cat single value, so its K was determined by competitive hydrolysis of mixed substrates with senescentin 1 peptide cat / K m Value (Fersht A "Emzyme Structure and Mechani...
Embodiment 3
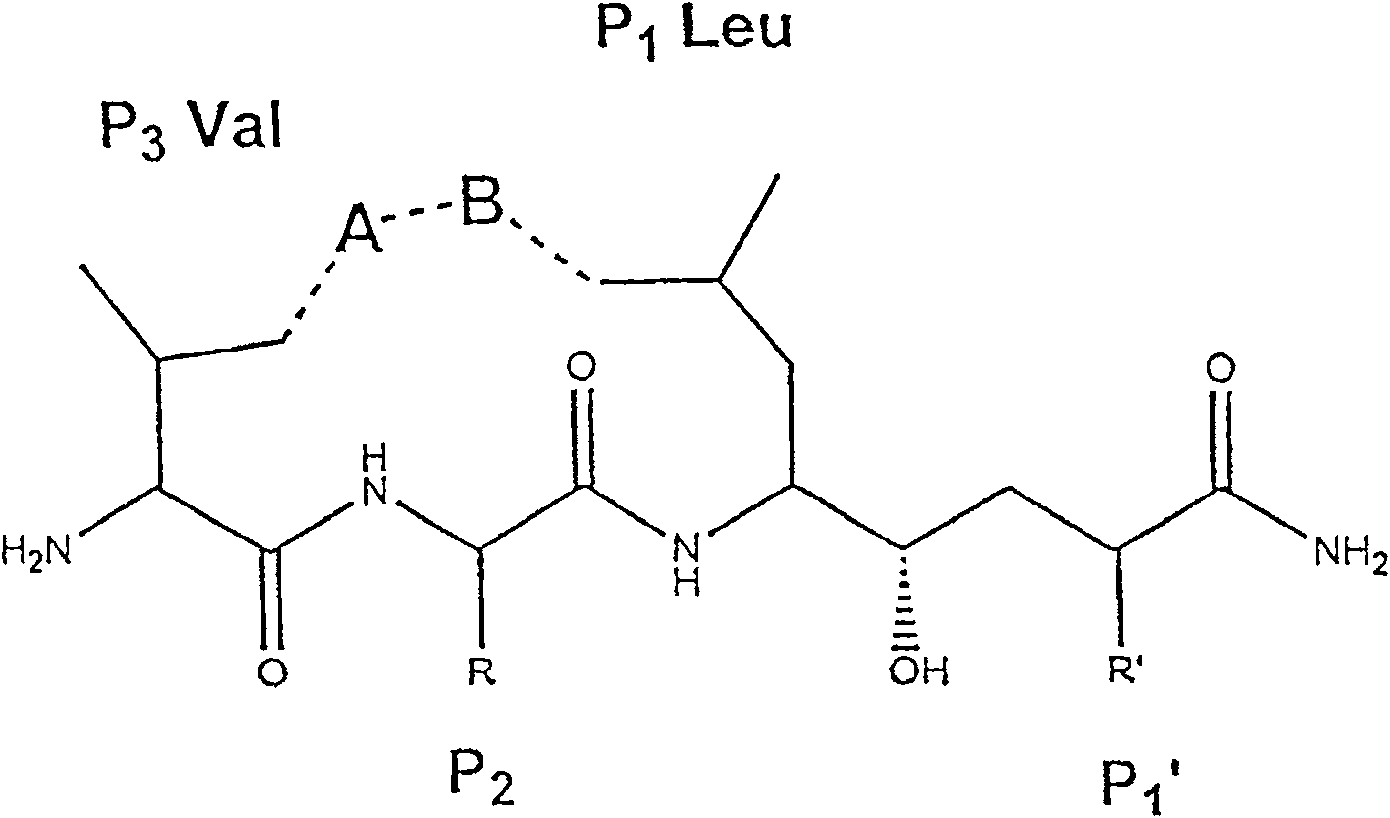
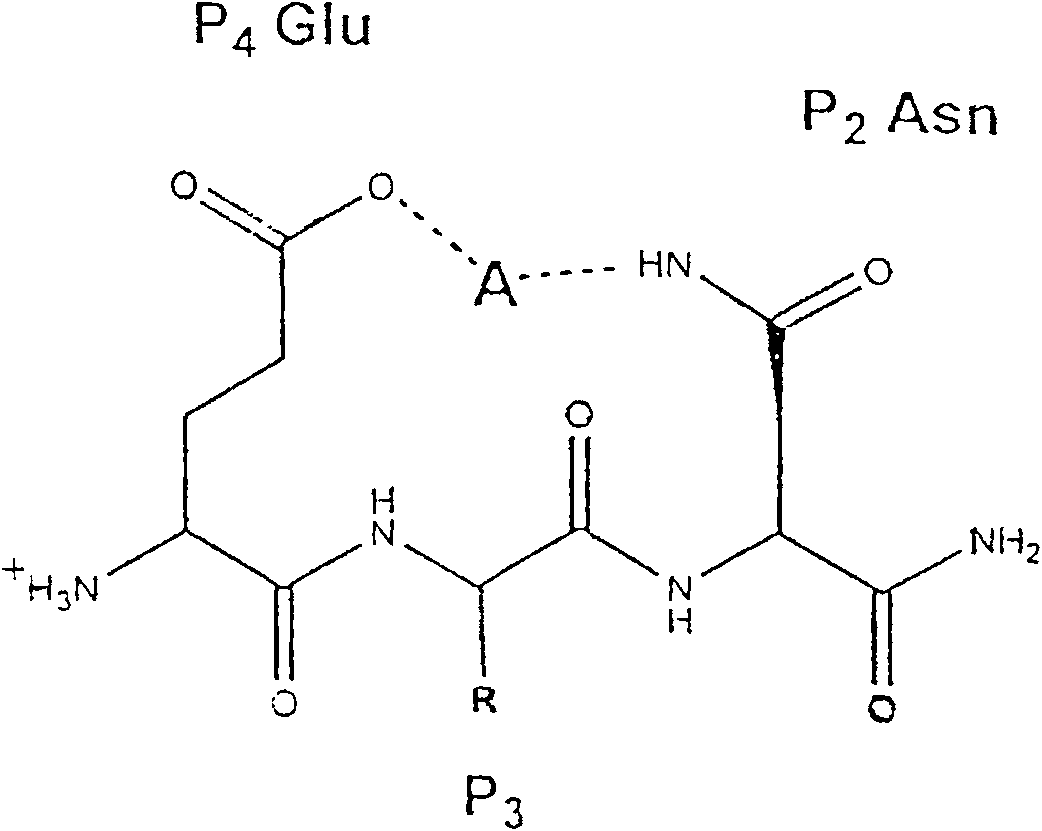
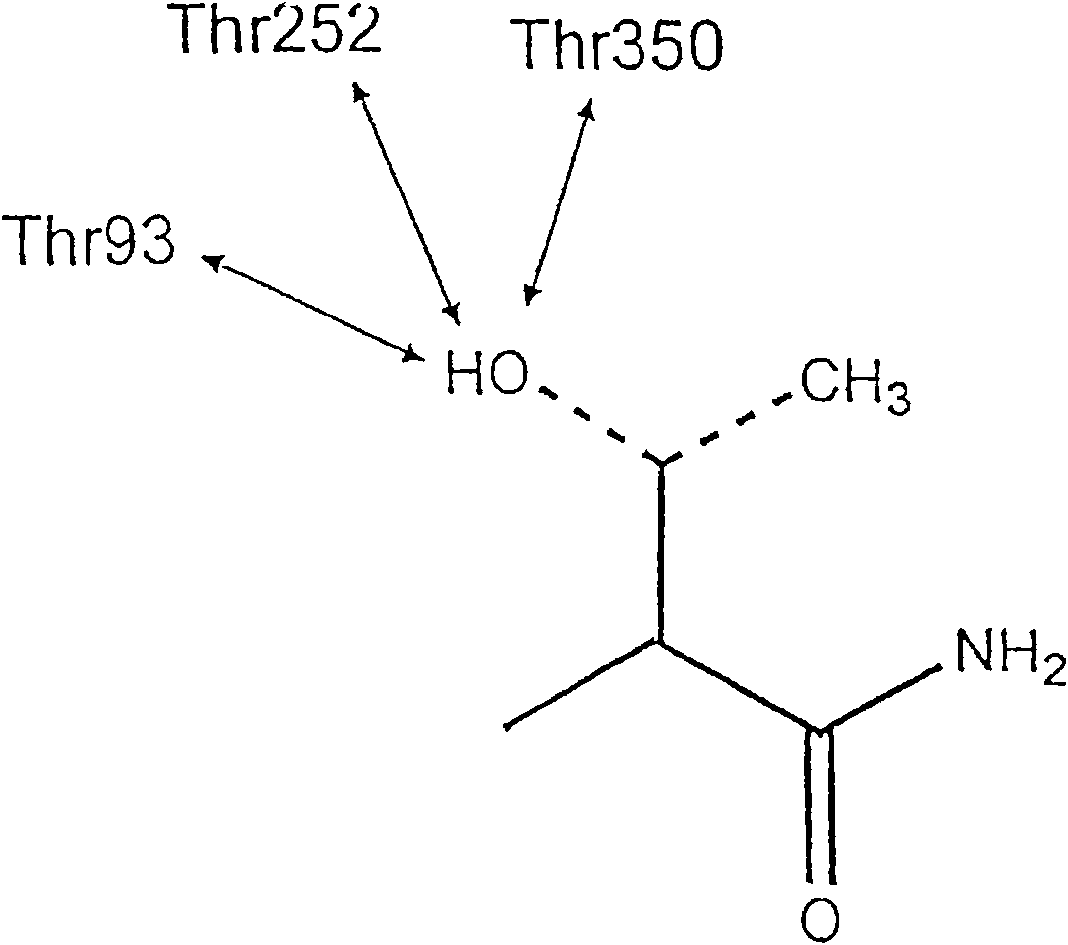
[0144] Example 3 Design and Synthesis of Memapsin2 Inhibitors OM99-1 and OM99-2
[0145] Based on the results of memapsin2 specificity, it was predicted that the residues suitable for P1 and P1' positions should be leucine and alanine. Next, as determined by the specificity data, P1' is preferably a small residue such as alanine and serine. However, the crystal structure (determined below) indicates that this site is also capable of accommodating a variety of large residues. It has been confirmed that P1' of memapsin2 is the most stringent position for specificity, and it is desirable to have small side chain residues, such as alanine, serine, aspartic acid. Alanine was selected for P1' mainly because its hydrophobicity is better than that of serine and aspartic acid, which is conducive to penetrating the blood-brain barrier and meets the design requirements of a memapsin2 inhibitor for the treatment of AD. Therefore, the inhibitor was designed to place a transition state ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com