Variant hypocrea jecorina cbh2 cellulases
A technology of H. jecorina and cellulase, applied in the direction of enzymes, enzymes, hydrolytic enzymes, etc., can solve the problems of low effectiveness
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0272] Alignment of known Cel6A cellulases
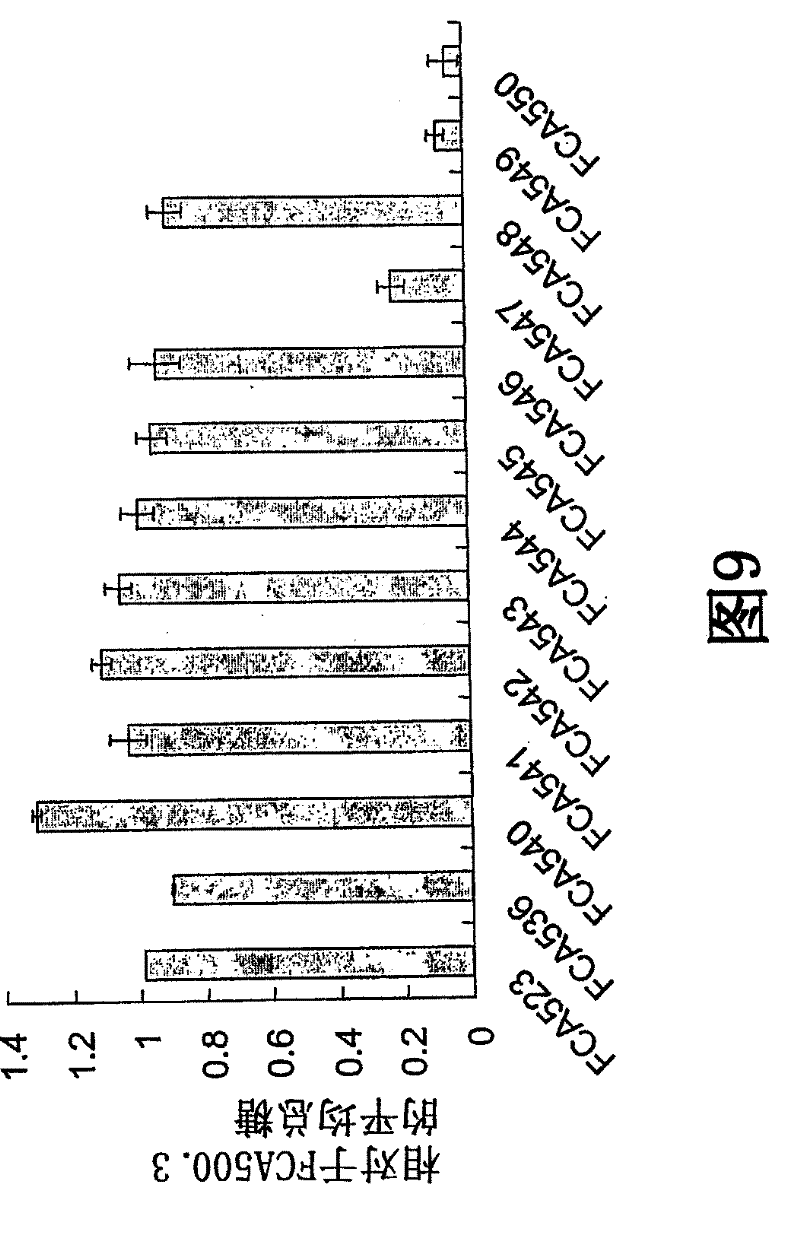
[0273] Selection for several mutations was determined by first aligning H. jecorina Cel6A to eight (8) family members using structural information and modeling programs. image 3 Derived from Humicola anomalies (Q9C1S9), Acremonium cellulotyticus (093837), Agaricus bisporus (P49075), Hypochaeta korningina (AF315681), Phanerochaete chrysosporium (S76141), Talaromyces emersonii (Q8N1B5 ), Lentinula edodes (AF244369), and H. jecorina (P07987) CBH2 molecular alignment. Using the VectorNTI Suite software program, the alignment was performed by Clustal W with a gap penalty of 10. [0233] Based on this alignment, various single and multiple amino acid mutations are made in the protein by site mutagenesis. By using the consensus sequence, possible mutations that might improve the thermostability of the enzyme were identified. see image 3 . Visual inspection of the 3D structures was performed to check their compatibility with the struct...
Embodiment 2
[0276] Preparation of cbh2 constructs
[0277] exist figure 2 The cDNA sequence of CBH2 described in was used as a template for the amplification of this gene. It also serves as a template for introducing mutations.
[0278]The following DNA primers were constructed for the amplification of mutant cbh2 genes from genomic DNA isolated from various microorganisms. All symbols used herein for protein and DNA sequences correspond to IUPACIUB Biochemical Nomenclature Commission codes.
[0279] Homologous 5' (FRG361 ) and 3' (FRG362) primers were developed based on the cbh2 sequence from T. reesei. Both primers contain Invitrogen The Gateway clone sequence. Primer 361 contains the attB1 sequence, while primer FRG362 contains the attB2 sequence.
[0280] FRG361 sequence without attB1:
[0281] ATGATTGTCGGCATTCTCAC (this initiates the 5' end of the gene encoding the signal sequence of H. jecorina CBH2) (SEQ ID NO: 3)
[0282] FRG362 sequence without attB2:
[0283] TTACAGGA...
Embodiment 3
[0297] Site Directed Mutagenesis
[0298] Based on the rationale described in Example 1, site-directed CBH2 mutants were prepared with the following 5' phosphorylation primers, which were developed and synthesized using techniques known in the art:
[0299] Table 3: Primers for single site-directed mutants
[0300] mutation Primer bps SEQ ID NO. V94E CAGGCAACCCTTTT GAA GGGGTCACTCCTTGGCG 34 P98L CAACCCTTTTGTTGGGGTCACT CTT TGGGCCAATGC 36 G118P GCTATTCCTAGCTTGACT CCA GCCATGGCCACTGCTG 37 M120L TCCTAGCTTGACTGGAGCC CTG GCCACTGCTGC 33 M134V GCTGTCGCAAAGGTTCCCTCTTT TGT GTGGCTAGATACTCTTG 43 T142V GATACTCTTTGACAAG GTC CCTCTCATGGAGCAAACCTTGGCC 42 M145L CAAGACCCCTCTC CTG GAGCAAACCTTGGCCGAC 34 T148Y GACCCCTCTCATGGAGCAA TAC TTGGCCGACATCCG 36
[0301] mutation Primer bps SEQ ID NO. T154A CGACATCCGCGCCGCCAACAAGAATGGCGG 30 L179A CGATTGCGC...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com