Method for calculating force between protein and DNA by computer simulation
A technology of analog computing and protein, applied in computing, special data processing applications, instruments, etc., to achieve the effects of easy preparation, high equipment utilization, and low equipment requirements
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
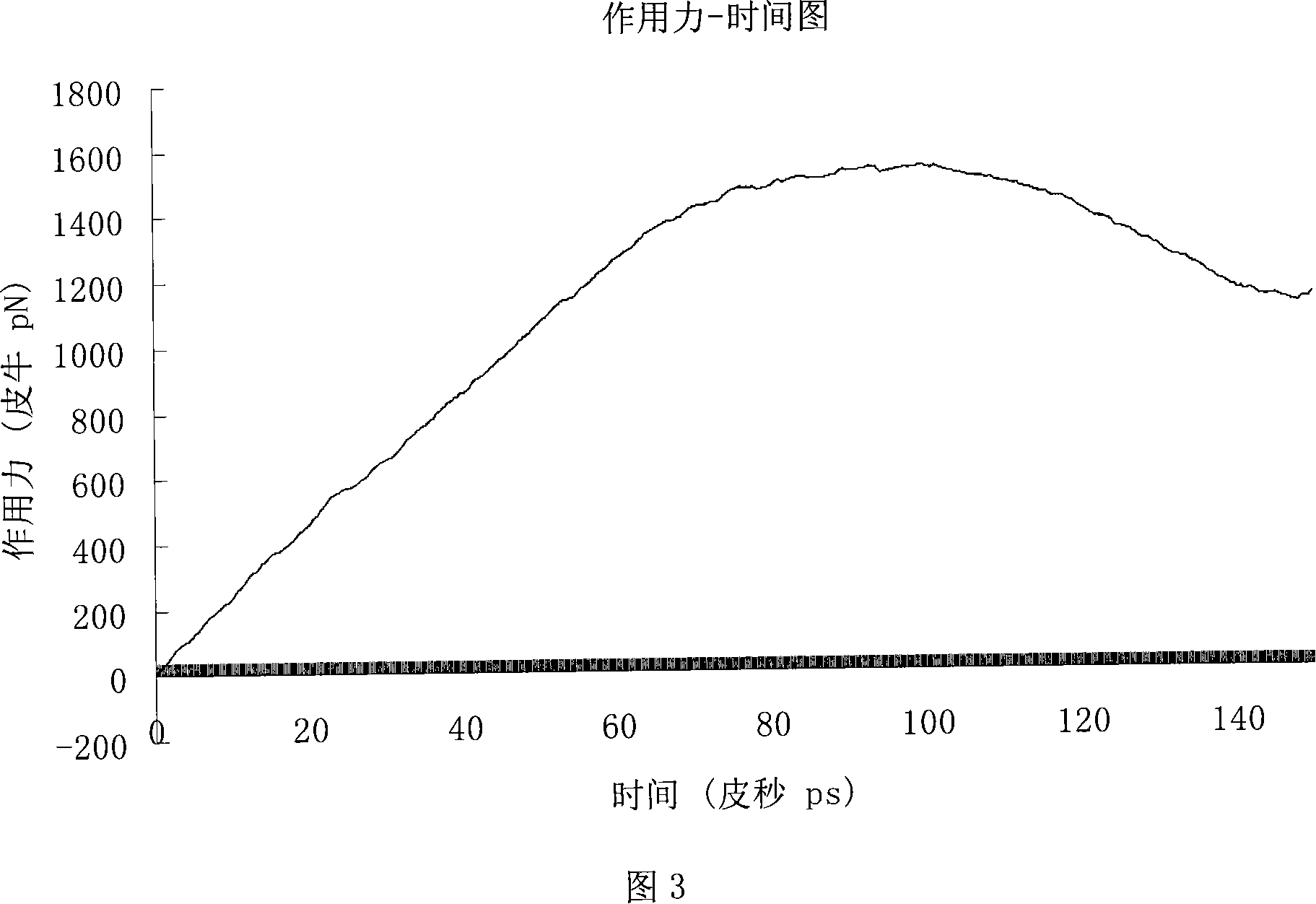
Image
Examples
Embodiment 1
[0035] (1) Obtain the complex data file coded 1o4x from the PDB database (http: / / www.rcsb.org / pdb / home / home.do), and name it 1o4x.pdb;
[0036] (2) Use VMD-1.8.6 software to extract the target protein and DNA fragments to be studied, and dissolve them into a cuboid water tank (84 Ȧ*70 Ȧ*61 Ȧ) filled with 0.9% NaCl solution to obtain protein-DNA- Salt solution system (see Figure 1); the structure containing non-hydrogen atom spatial position data in the system is named 1o4x_ionized.pdb, and the structure containing hydrogen atoms and hydrogen bond parameters is named 1o4x_ionized.psf;
[0037](3) Utilize NAMD_2.6 software, adopt Charmm27 force field, use Steepest Descent method, under periodic boundary conditions (Periodic Boundary Conditions PBC), the total time length of the above-mentioned protein-DNA-salt solution system is 100 picoseconds (ps) ) energy minimization (Minimize) process, in which every 1 femtosecond (fs) calculates the van der Waals force of other atoms in th...
Embodiment 2
[0046] (1) Obtain the complex data file coded 1wtq from the PDB database (http: / / www.rcsb.org / pdb / home / home.do), named 1wtq.pdb;
[0047] (2) Utilize the VMD-1.8.6 software to extract the target protein and DNA fragments to be studied, and dissolve them in a cuboid water tank filled with 0.9% NaCl solution to obtain a protein-DNA-salt solution system; the system contains non- The structure of the hydrogen atom spatial position data is named 1wtq_ionized.pdb, and the structure containing hydrogen atoms and hydrogen bond parameters is named 1wtq_ionized.psf;
[0048] (3) Utilize NAMD_2.6 software, adopt Charmm27 force field, use Steepest Descent method, under periodic boundary conditions (Periodic Boundary Conditions PBC), the total time length of the above-mentioned protein-DNA-salt solution system is 100 picoseconds (ps) ) energy minimization (Minimize) process, in which every 1 femtosecond (fs) calculates the van der Waals force of other atoms in the spherical space with each...
Embodiment 3
[0057] (1) Obtain the complex data file coded 1hry from the PDB database (http: / / www.rcsb.org / pdb / home / home.do), named 1hry.pdb;
[0058] (2) Utilize the VMD-1.8.6 software to extract the target protein and DNA fragments to be studied, and dissolve them in a cuboid water tank filled with 0.9% NaCl solution to obtain a protein-DNA-salt solution system; the system contains non- The structure of the hydrogen atom spatial position data is named 1wtq_ionized.pdb, and the structure containing hydrogen atoms and hydrogen bond parameters is named 1wtq_ionized.psf;
[0059] (3) Utilize NAMD_2.6 software, adopt Charmm27 force field, use Steepest Descent method, under periodic boundary conditions (Periodic Boundary Conditions PBC), the total time length of the above-mentioned protein-DNA-salt solution system is 100 picoseconds (ps) ) energy minimization (Minimize) process, in this process, every 1 femtosecond (fs) calculates the van der Waals force of other atoms in the spherical space w...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com