Nucleotides sequence, molecule probe and method for identifying zhejiang fritillary variant-dong fritillary
A technology of nucleotide sequence and nucleic acid molecules, which is applied in the field of identification of Fritillaria varietata---East Fritillary. It can solve the problems of research reports and unseen technology for fast identification of Fritillaria fritillaria DNA, and achieve less sample consumption. Effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0040] Example 1: Preparation of Orient Fritillaria Specific Nucleotide Sequence
[0041] 1 Genomic DNA extraction
[0042] The modified CTAB method (Doyle, 1991) was used to extract genomic DNA from fritillaria leaves, and the steps were as follows:
[0043] (1) Take 0.05 g of silica gel-dried Fritillaria leaf material, put it in a BIO101 tube and grind it into powder, then add 2ml of 2×CTAB extraction buffer (100mM Tris-HCL (PH8.0), 20mM EDTA, preheated at 65°C, 1.4M NaCl, 2% CTAB, 2% β-mercaptoethanol), gently shake to disperse the solution evenly, 65°C water bath for 20-30 minutes, gently shake every 5 minutes during the water bath.
[0044] (2) Cool to room temperature, add an equal volume of chloroform:isoamyl alcohol (24:1), mix well, and centrifuge at 10,000 g for 10 minutes.
[0045] (3) Transfer the upper aqueous phase into a new 5ml centrifuge tube, add 1 / 10 volume of 10×CTAB-0.7MNaCl, mix gently, then add an equal volume of chloroform:isoamyl alcohol (24:1) for e...
Embodiment 2
[0064] Embodiment 2: Preparation of friD1 and friD2 specific nucleic acid molecular probes of Fritillaria fritillaria
[0065] On the basis of obtaining the specific nucleotide sequence of Fritillaria fritillaria, use Primer Primer 5.0 (VinaySingh, 1998) software to design, draw friD1 and friD2 (respectively 79-97bp and 661 bp shown in 1 in the sequence list The nucleotide composition and arrangement of -679bp) are good oligonucleotide fragments for identification of Fritillaria japonica. According to the nucleotide composition arrangement of friD1 and friD2 (the nucleotide composition and arrangement shown in the 2 and 3 sequences in the sequence listing), they were synthesized on an automatic DNA synthesizer.
Embodiment 3
[0066] Embodiment 3: the identification (conventional PCR method) of Eastern Fritillaria
[0067] 1. Extraction of DNA: using the improved CTAB method to extract DNA from Fritillaria
[0068] 2. The PCR reaction system is:
[0069] 10×Buffer(Mg 2+ free)
MgCl 2 (25mM)
dNTPs (10mM)
friD1 (5 μM)
FriD2 (5μM)
Taq enzyme (5U / μl)
Template DNA (30ng / μl)
ddH2O
2.5μl (1×Buffer)
2μl (2.0mM)
0.5μl (0.2mM)
1μl (0.4μM)
1μl (0.4μM)
0.25μl (1.2U)
2.5μl (60ng)
15.76μl
total capacity
25μl
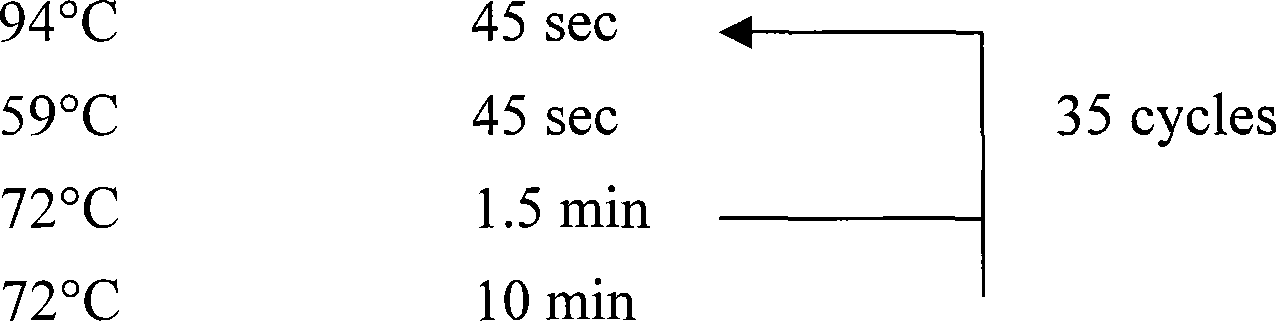
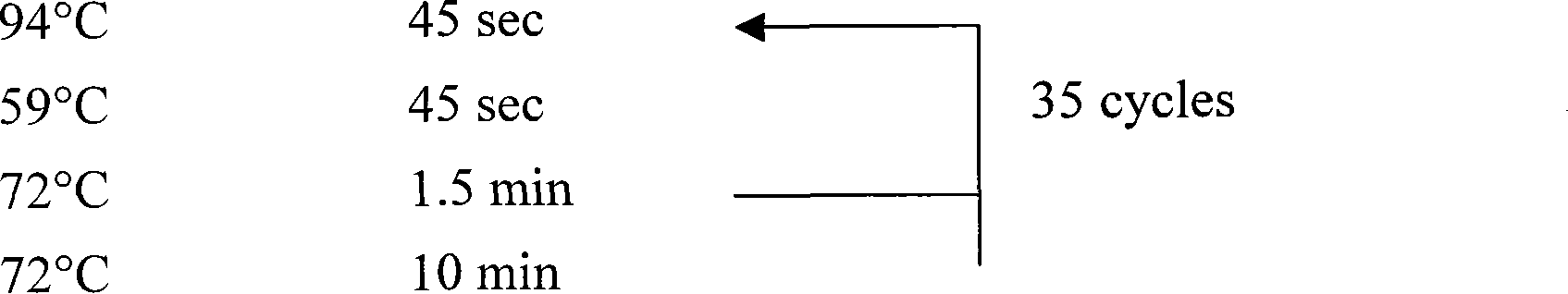
[0070] 3. PCR operation: Take two 0.5 ml PCR tubes, add 22.5 microliters of PCR mixture according to step 2, then add 2.5 microliters of DNA to one tube, and 2.5 microliters of PCR mixture (control) into the other tube, and put On the PCR instrument, carry out the PCR reaction according to the following procedure:
[0071] 94℃ 5min
[0072]
[0073] The results of PCR amplification we...
PUM
| Property | Measurement | Unit |
|---|---|---|
| Molecular weight | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com