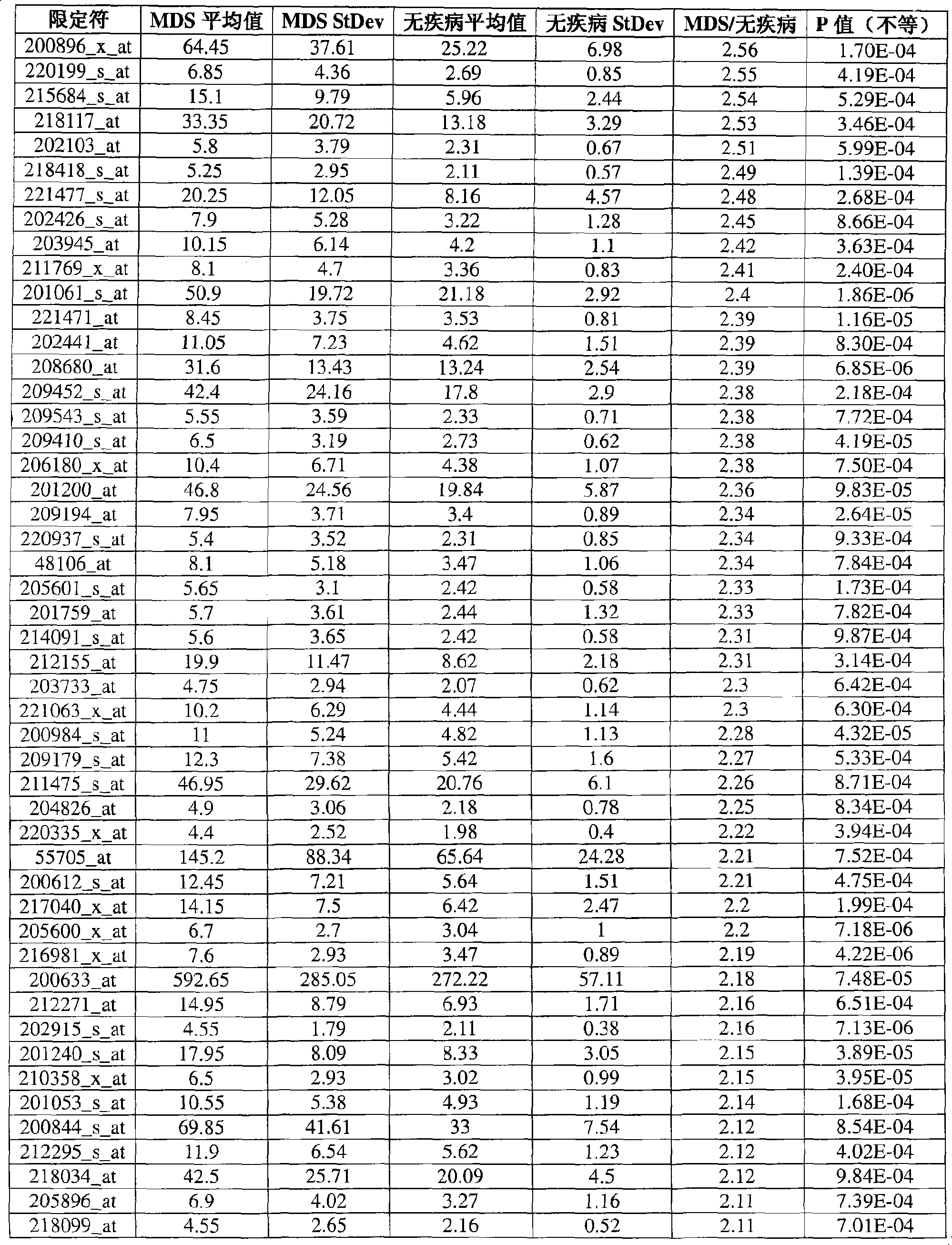
Leukemia disease genes and uses thereof
A leukemia and gene technology, applied in the field of leukemia disease genes and the use of said leukemia disease genes to diagnose and treat leukemia, can solve laborious and time-consuming problems
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
example 1
[0180] Example 1. Purification of PBMC and RNA
[0181] All blood was collected from diseases from disease volunteers, MDS patients and AML patients. Receive the awareness of the drug dynamics part of these clinical studies and enable the scheme to be approved by the local human test committee (INSTITIONAL REVIEW BOARD) at the clinical site. MDS patients are mainly a caucasian and an average age of 66 years (within 52-84 years). AML patients are only Caucasian and have an average age of 45 (within 19-65 years). Disease-free volunteers are only a caucasian and have an average age of 23 years (within 18-32 years).
[0182]The inclusion standards of AML patients include 20% of the bone marrow, and the morphological diagnosis of AML performed by the FAB classification system; and instructions CD33 + State flow cytometry. The inclusion criteria of MDS patients include MDS morphological diagnosis, and refractory anemia, refractory anemia accompanied by cyclic ferrochromic cells, materna...
example 2
[0184] Example 2. RNA amplification and GeneChip Generation of hybridization probes.
[0185] Use the modified Lockhart et al. (1996) Nature Biotechnology The procedures described in 14: 1675-1680 are used to prepare a labeled target for oligonucleotide arrays. 2 micrograms total RNA were converted to cDNA using oligo-(DT) 24 primers containing T7DNA polymerase promoters. The cDNA was used as a template using a T7DNA polymerase kit (Ambion, Woodlands, TX, USA) and biotinylated CTP and UTP (Enzo, Farmingdale, NY, USA). At 94 ° C, the labeled cRNA was broken for 35 minutes in 40 μl of the final volume of 40 mM Tris-acetate (pH 8.0), 100 mM KOAC, 30 mM mgOAc.
example 3
[0186] Example 3. Hybridization and fluorescence detection with AFFYMETRIX microarray
[0187] Individual illness and disease-free samples with HG-U133a GeneChips (Affymetrix) hybridization. No samples are collected. The tagged target was diluted in 1 × MES buffer having 100 μg / ml of herring DNA and 50 μg / mL acetylation BSA. Such like Hill et al. (2000) Science The arrays are standardized to each other and to evaluate the sensitivity of the oligonucleotide array, such that each hybridization reaction includes 11 bacterial genes in vivo synthesis transcripts. The abundance of these transcripts is within the range of 1: 300,000 (3 ppm) to 1: 1000 (1000 ppm) on the number of control transcripts on the overall transcript. The detection sensitivity of the array can be within the range of from about 1: 300,000 and 1: 100,000 copies of the signal by the signal reactivity of these control transcripts.
[0188] The labeled probe was denatured at 99 ° C for 5 minutes, and then denatur...
PUM
 Login to view more
Login to view more Abstract
Description
Claims
Application Information
 Login to view more
Login to view more - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap